Accessibility of HTML pages from R Markdown Tweet
Accessibility
Over the years, I faced situations where I needed to make content (lecture notes, manuscripts, consultancy reports, etc.) as accessible as possible to effectively communicate with colleagues, clients, and students with particular accessibility requirements.
All those “accessibility encounters” of mine left me with a pile of unstructured notes and advice to my self to which I often refer. Well, I decided to blog about parts of it in case anyone else finds it useful.
Given how pervasive R Markdown is getting when it comes to authoring and exchanging content in the Data Science ecosystem (and will get even more pervasive now that Python integrates so nicely with R via the reticulate R package), in this short blog post, I want to focus on simple steps to increase the accessibility of HTML pages generated using R Markdown.
Other people have dealt with aspects of the problem before (see External links and resources below for a few links), so I do not claim that there is anything particularly original or novel in the content here. What is perhaps useful to me (and hopefully to others!) is having it all in one place. Also, bear in mind that I neither have nor claim any particular accessibility expertise; all I have to talk about is a few happy colleagues, students, and clients over the years.
Headings
It is all too tempting not to have any or not to care about the hierarchy of headings in an R Markdown file, especially since the default is to show no numbering and no indication of hierarchy other than the heading font size.
For a sighted reader, no hierarchy or no numbering is usually not an issue; the order of the text blocks is typically sufficient for them to understand the flow of content and construct a mental map of the hierarchy.
However, when the content is accessed using a screen reader, the situation is different. Most screen readers will speak out the level of each heading before reading out the heading text.
For example, imagine how confusing it can be for a first-year statistics student if they were listening to a screen reader reading off the table of contents of their basic probability lecture notes like:
- “Heading level one: Introduction to probability”
- “Heading level three: The history of probability”
- “Heading level two: Interpretations of probability”
- “Heading level one: Experiments and events”
- “Heading level three: Set theory”
- “Heading level two: The definition of probability”
Since, by default, the title of an R Markdown document (what is in the
YAML content) ends up being heading of top level in the HTML output
(heading level 1 typically), I recommend starting with ## for
sections, ### for the sub-sections, etc.
URLs and hyperlinks
Screen readers work better with hyperlinked text than plain URLs or paths. For example, the following Markdown text
> [Color Universal Design](https://jfly.uni-koeln.de/color/) is better than just <https://jfly.uni-koeln.de/color/>in a browser looks like
Color Universal Design is better than just https://jfly.uni-koeln.de/color/
which is read by macOS’ screen reader (press CMD+F5 to enable) like “Link Color Universal Design is better than just link h t t p s colon slash slash accessibility dot j fly dot uni dash …”. You get the picture (or better the sound).
The Accessible hyperlinks tutorial by NC State University provides more advice on setting up accessible hyperlinks.
Code output
It is best to avoid the default behaviour of using leading hashes (##) in the text output of code chunks, or any other fancy output indicators you may be using. The reason is that leading hashes are read as “number number” by a screen reader, which can get tedious with long output. You can bypass the default behaviour by setting the chunk option comment = "".
To illustrate, the code chunk
```{r}
a <- 3
a
```gives
a <- 3
a## [1] 3Instead, the code chunk
```{r, comment = ""}
a <- 3
a
```gives
a <- 3
a[1] 3Alternatively, you can add the code chunk
```{r, echo = FALSE}
library("knitr")
opts_chunk$set(comment = "")
```at the top of you Rmd file (or add comment = "" to your options if
you are already setting other options), to set comment = "" to be
the deafult for all code chunks in the R Markdown file.
Alt text, titles and captions
Images from external files
The <img> HTML tag has an alt attribute, which is what a screen reader will read when it gets to the image. This is our opportunity to provide some alt text for readers relying on screen readers to access the content. The <img> HTML tag also has a title attribute that is shown as a tooltip when you hover over the image in a browser.
Markdown’s  syntax for including images from external files allows for the inclusion of all alt text, title text, and captions. However, depending on whether the image is in a paragraph or whether A-text or B-text are supplied or not, what ends up being the title, caption, and alt text varies. Of course, it varies in an entirely predictable way, but still, it can get hard to remember to break that line or add a \ after the inclusion of an image (at least for me).
R Markdown’s default behaviour, on the other hand, is to use the text for a figure caption, both as a caption and as alt text, which is not ideal in many circumstances.
For those reasons, when I author R Markdown documents I rely on knitr::include_graphics() and the custom knitr hook below that allows me to specify any or all of the title, alt-text and caption.
my_plot_hook <- function(x, options) {
base <- knitr::opts_knit$get('base.url')
if (is.null(base)) {
base <- ""
}
alt <- ifelse(is.null(options$alt), "", options$alt)
cap <- ifelse(is.null(options$caption), "", options$caption)
tit <- ifelse(is.null(options$title), "", options$title)
w <- options[['out.width']]
h <- options[['out.height']]
if (!is.null(w) | !is.null(h)) {
str <- paste(c(sprintf('width=%s', w),
sprintf('height=%s', h)), collapse = " ")
str <- paste("{", str, "}")
}
if (cap == "") {
sprintf('%s\\', alt, base, x, paste0("'", tit, "'"), str)
}
else {
sprintf('%s\\\n\n<blockquote><p class = "caption">%s</p></blockquote>', alt, base, x, paste0("'", tit, "'"), str, cap)
}
}The above hook is an extended version of a hook that appeared in a discussion at RStudio’s community forum. It allows us to specify alt text, caption text (within a <blockquote> HTML tag to distinguish it from the standard text) and title text, all using chunk options. Before I continue with a demo, note that the hook is specific to getting HTML output from a single R Markdown file via pandoc. I cannot provide any guarantees of it working as advertised for other output formats or more involved collections of R Markdown files (e.g. bookdown projects). See, for example, knitr’s GitHub repository for how involved the code for the default plot hooks has to be to deliver support for a range of output formats. Also, any other figure- and image-specific chunk options other than title, alt, caption, out.width, and out.height will be ineffective when you are using this hook (but that’s all the options I typically need with HTML content).
Let’s replace the default plot hook with my_plot_hook
library("knitr")
knit_hooks$set(plot = my_plot_hook)Then, the code chunks
``{r, echo = FALSE}
cap_txt <- "Grey cat. A picture by Thomas Gass downloaded from [Wikimedia](https://commons.wikimedia.org/wiki/Category:Felis_silvestris_catus#/media/File:Greycat.jpg) distributed under [CC BY 2.5](https://creativecommons.org/licenses/by/2.5)."
alt_txt <- "'Misty', female cat, aged around two and a half years."
title_txt <- "A grey cat staring at you."
img_url <- "https://upload.wikimedia.org/wikipedia/commons/1/15/Greycat.jpg"
```
```{r, echo = FALSE, caption = cap_txt, alt = alt_txt, title = title_txt}
knitr::include_graphics(img_url)
```give

Grey cat. A picture by Thomas Gass downloaded from Wikimedia distributed under CC BY 2.5.
If you hover over the image (but do not to look into the cat’s eyes for too long!) you get a tooltip with the title “A grey cat staring at you”. The caption is the quoted text immediately below the image, and a screen reader will say “Misty, female cat, aged around two and a half years” when the image is selected.
Images generated by code-blocks
We can use the same hook for images that are generated programmatically. For example, suppose that we want to demonstrate the color-blind friendly palettes recommended by the Color Universal Design using a bar plot. We first create the character strings with the alt text and title text we want to have (just to keep the chunk options tidy).
alt_txt <- "A barplot using the color-blind friendly palettes recommended by [Color Universal Design](https://jfly.uni-koeln.de/color/)."
title_txt <- "A colorblind-friendly palette"Then, the following chunk of code
```{r, title = title_txt, alt = alt_txt}
cbPalette <- c("#999999", "#E69F00", "#56B4E9", "#009E73", "#F0E442", "#0072B2", "#D55E00", "#CC79A7")
barplot(rep(1, length(cbPalette)), col = cbPalette)
```returns
cbPalette <- c("#999999", "#E69F00", "#56B4E9", "#009E73", "#F0E442", "#0072B2", "#D55E00", "#CC79A7")
barplot(rep(1, length(cbPalette)), col = cbPalette)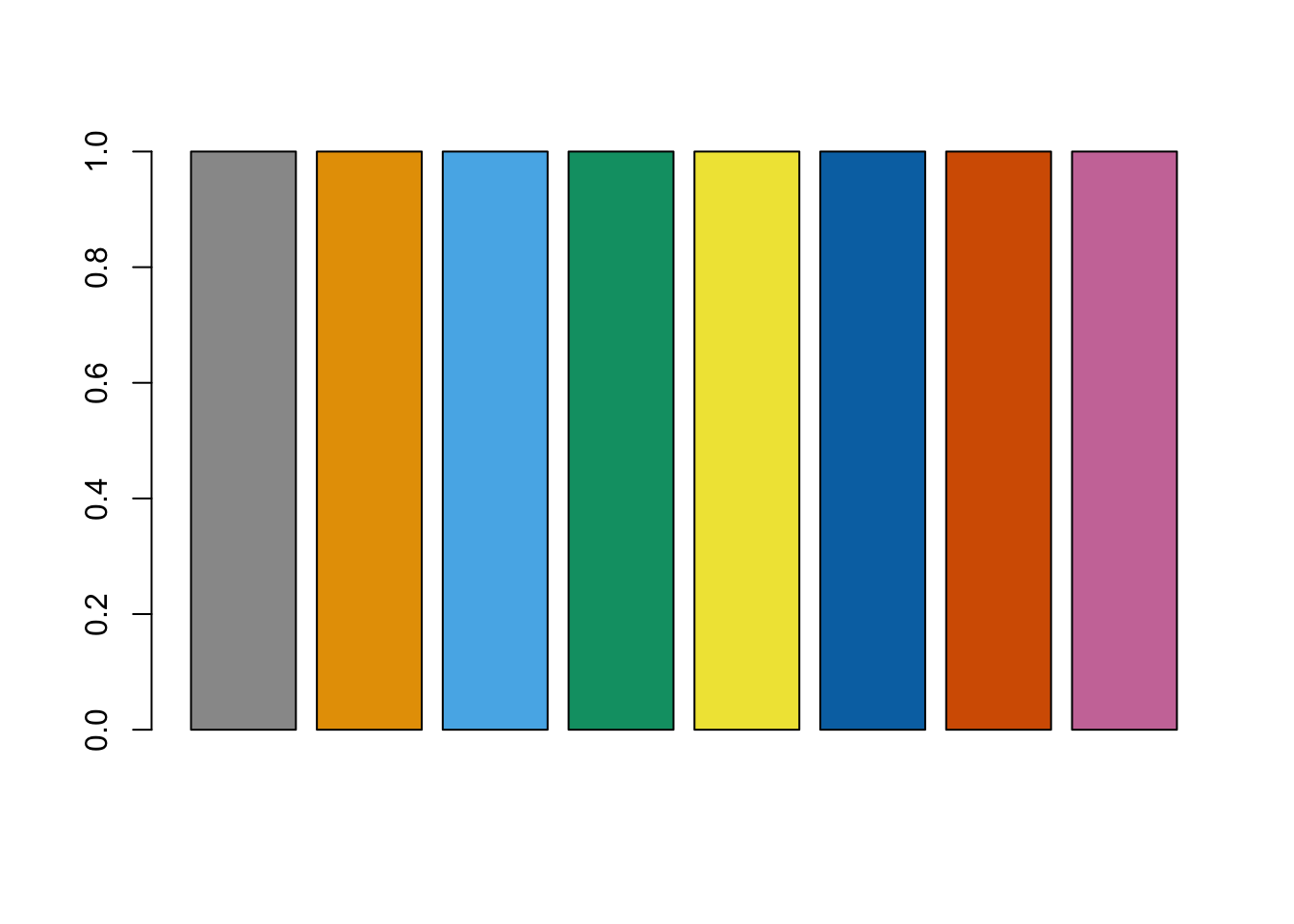
Palettes for color vision deficiencies
One of the main accessibility issues I see on web-pages (including my own) and books is the use of colors that are not friendly to readers with color vision deficiencies. The links and resources below can help you improve your pages, documents and graphics to cater as much as possible for such deficiencies.
colorbrewer2.org
colorbrewer2.org provides an easy-to-use dashboard to select palettes, with the option to limit only to colourblind-friendly ones.
RColorBrewer R package
The RColorBrewer R package can be used to generate colourblind-friendly palettes (in the form of vectors of hex colour codes). For example,
library("RColorBrewer")
display.brewer.all(colorblindFriendly = TRUE)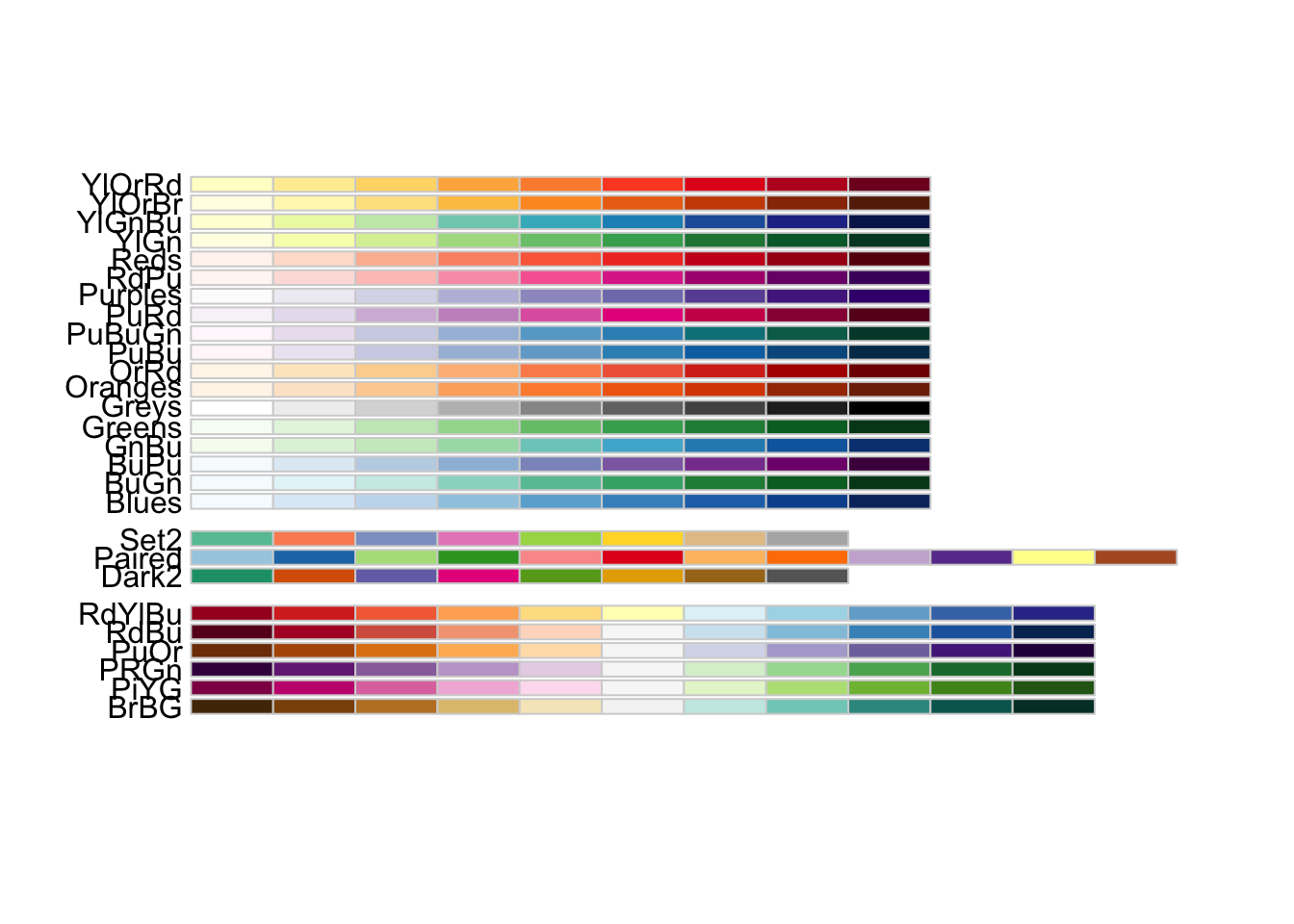
RColorBrewer’s colorblind friendly palettes.
PuBuGn <- brewer.pal(n = 9, name = "PuBuGn")
PuBuGn[1] "#FFF7FB" "#ECE2F0" "#D0D1E6" "#A6BDDB" "#67A9CF" "#3690C0" "#02818A"
[8] "#016C59" "#014636"barplot(rep(1, length(PuBuGn)), col = PuBuGn)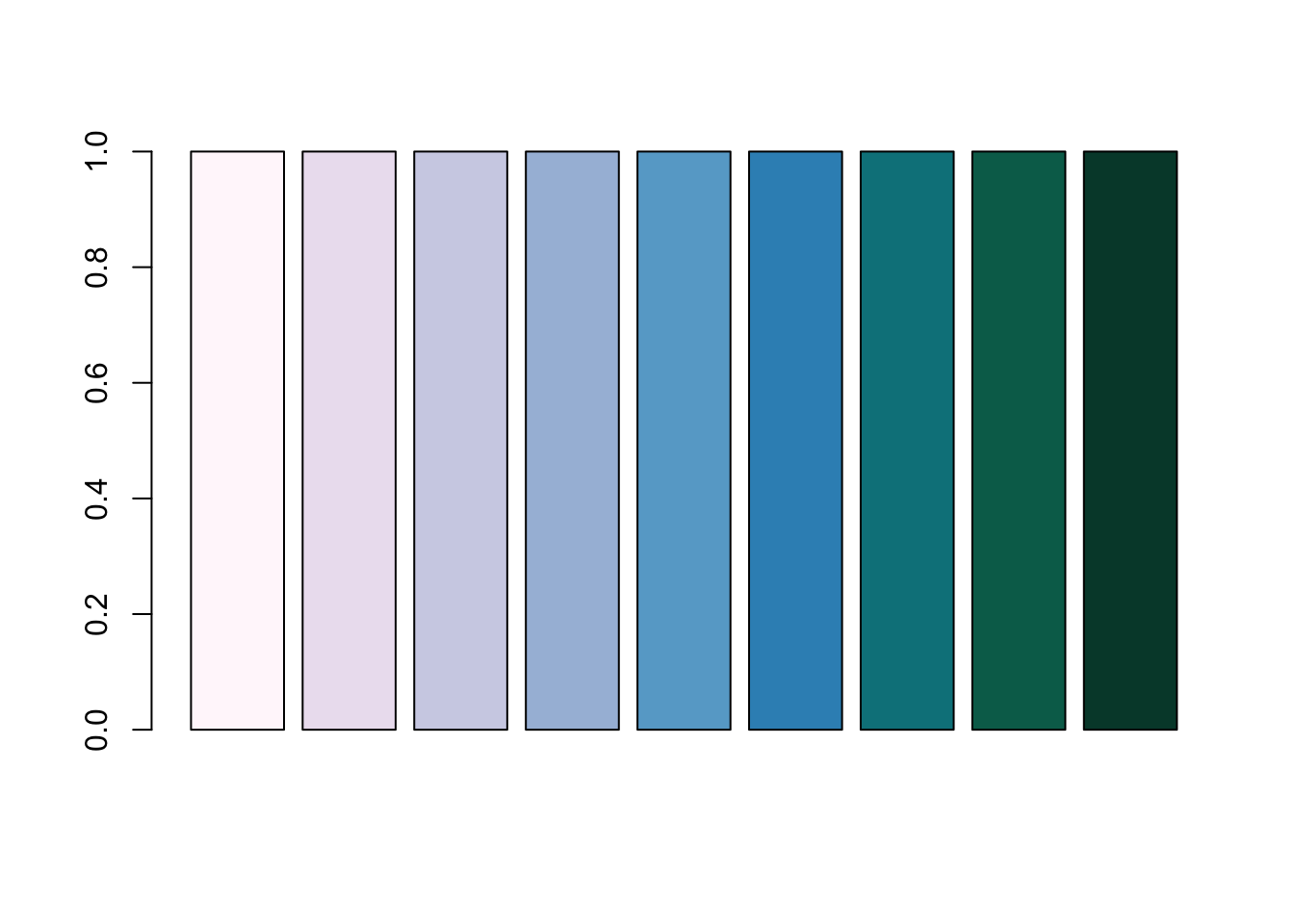
A barplot demonstating the RColorBrewer’s PuBuGn palette.
colorspace R package
The colorspace R package is a complex tool with a simple interface whose core functionality is to “carry out mapping between assorted color spaces including RGB, HSV, HLS, CIEXYZ, CIELUV, HCL (polar CIELUV), CIELAB and polar CIELAB” (directly from the package description). It also provides something handy: an emulator of color vision defficiencies both through the command line and colorspace’s GUIs. See the color vision deficiency emulation article from the colorspace web pages.
Accessing the colospace shiny GUI is as simple as
library("colorspace")
hcl_wizard()On the GUI, you can choose to see how the colors look like to someone
with normal, deutan (green-blindness), protan (red-blindness) and
tritan (blue-blindness) vision, or even wholly desaturate the
colours to see if you can distinguish them from each other. The
functions deutan(), protan(), and tritan() take as input a
vector of colors and return the hex color codes of how that colour
looks like depending on the severity of each vision deficiency. For
example,
severity <- seq(0, 1, 0.1)
protan_greens <- sapply(severity, protan, col = "green")
barplot(rep(1, length(severity)), col = protan_greens, xlab = "severity of protanopia")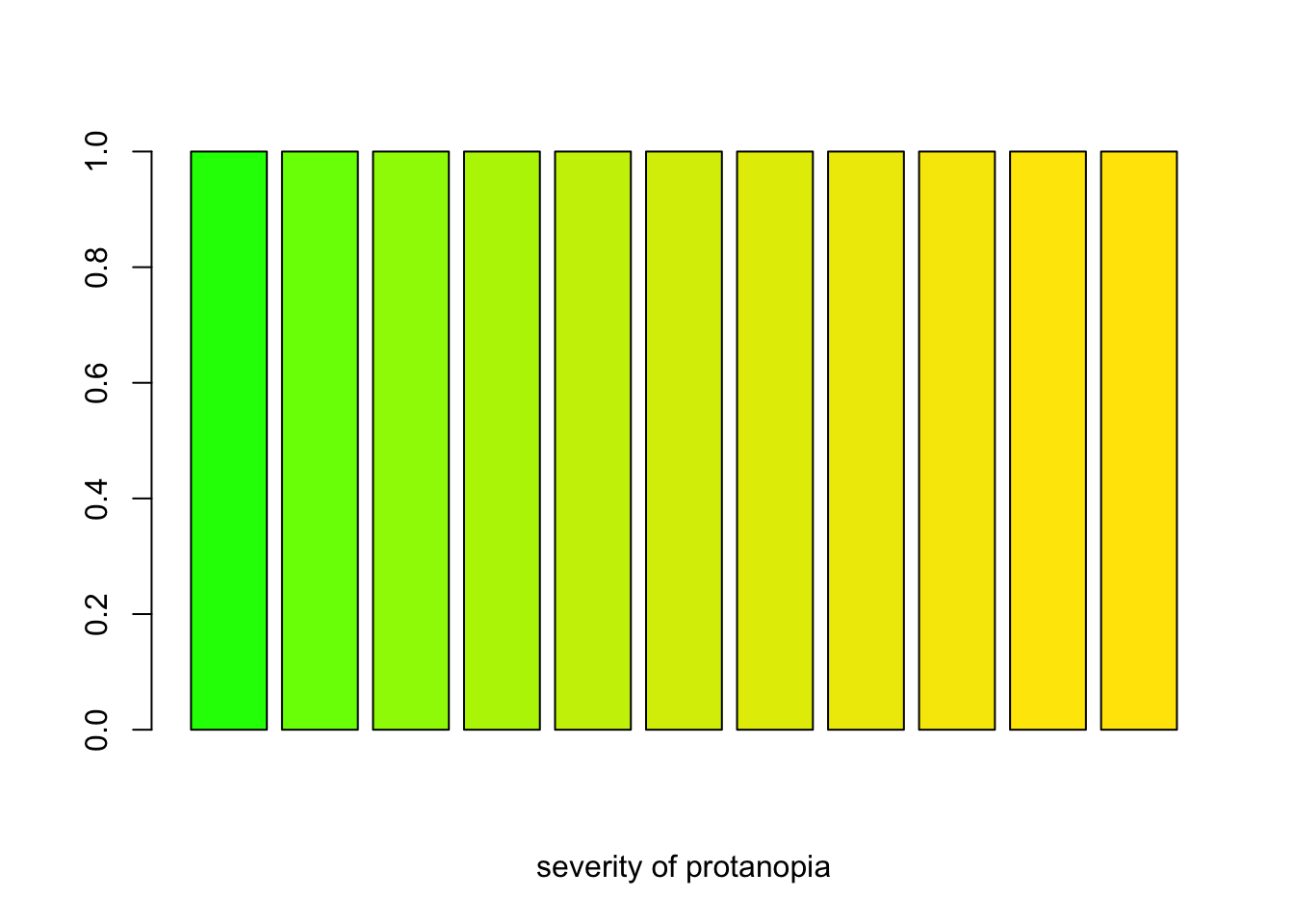
A barplot demosntrating how R’s color ‘green’ looks like for varying severity of protanopia.
viridis R package
The palettes provided by the
viridis R package
are designed to be “perfectly perceptually-uniform, both in regular
form and also when converted to black-and-white. They are also
designed to be perceived by readers with the most common form of color
blindness (all color maps in this package) and color vision deficiency
(‘cividis’ only)” (directly from the package description). For
example, the often-encountered rainbow palette scores very low when
displaying information to individuals with color deficiencies:
rain <- rainbow(n = 30)
par(mfrow = c(2, 2), mar = c(2, 2, 2, 2))
barplot(rep(1, length(rain)), col = rain, main = "no deficiency")
barplot(rep(1, length(rain)), col = protan(rain), main = "protanopia (red-blindness)")
barplot(rep(1, length(rain)), col = deutan(rain), main = "deutanopia (green-blindness)")
barplot(rep(1, length(rain)), col = tritan(rain), main = "tritanopia (blue-blindness)")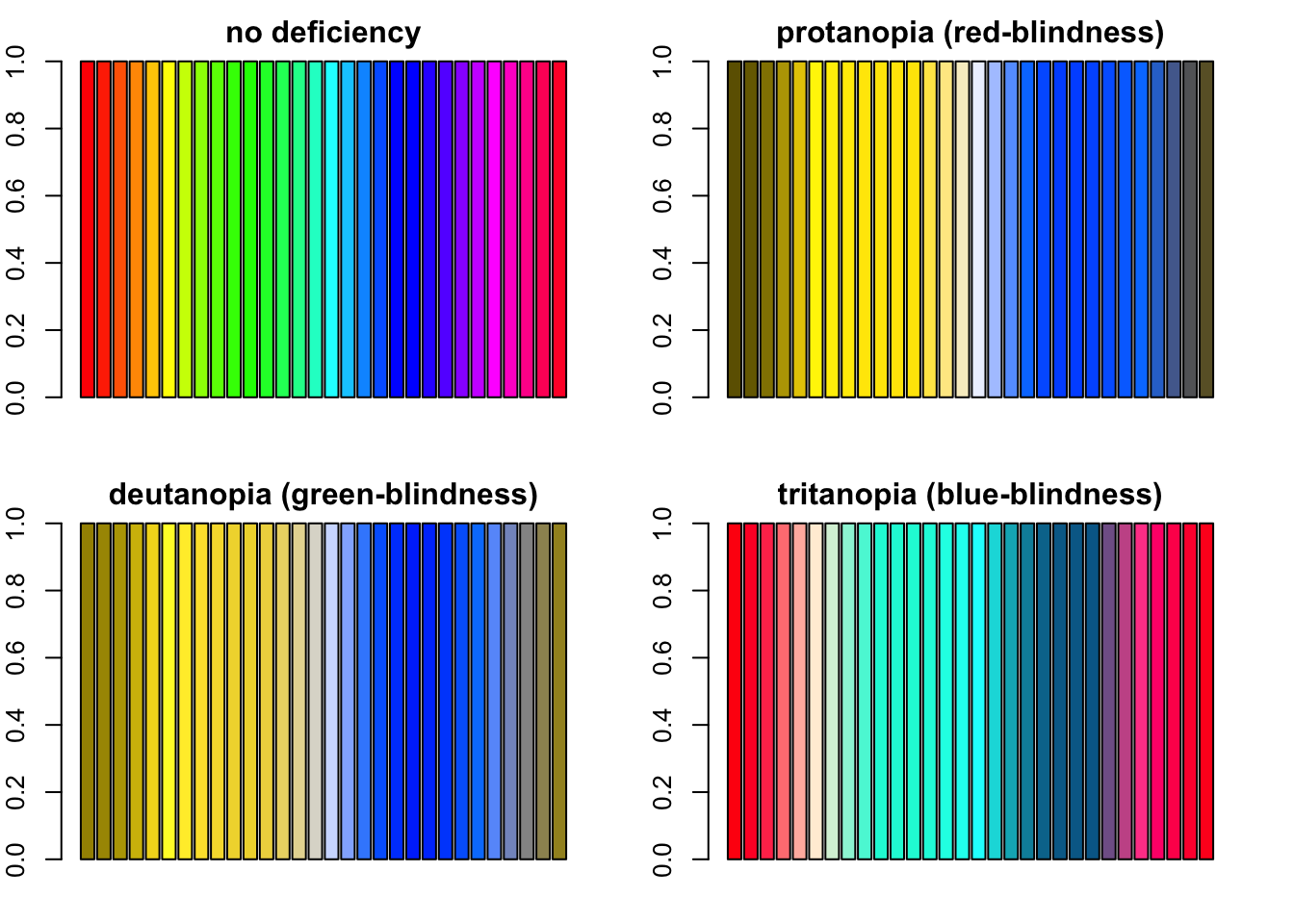
Four barplots demonstating how R’s rainbow palette looks like with severe protanopia, deutanopia and tritanopia.
On the other hand, viridis’ inferno works well (you should see a
sequence of 30 bars from darker to lighter regardless of which of the
four barplots you are looking at).
library("viridis")Loading required package: viridisLiteinferno <- inferno(n = 30)
par(mfrow = c(2, 2), mar = c(2, 2, 2, 2))
barplot(rep(1, length(inferno)), col = inferno, main = "no deficiency")
barplot(rep(1, length(inferno)), col = protan(inferno), main = "protanopia (red-blindness)")
barplot(rep(1, length(inferno)), col = deutan(inferno), main = "deutanopia (green-blindness)")
barplot(rep(1, length(inferno)), col = tritan(inferno), main = "tritanopia (blue-blindness)")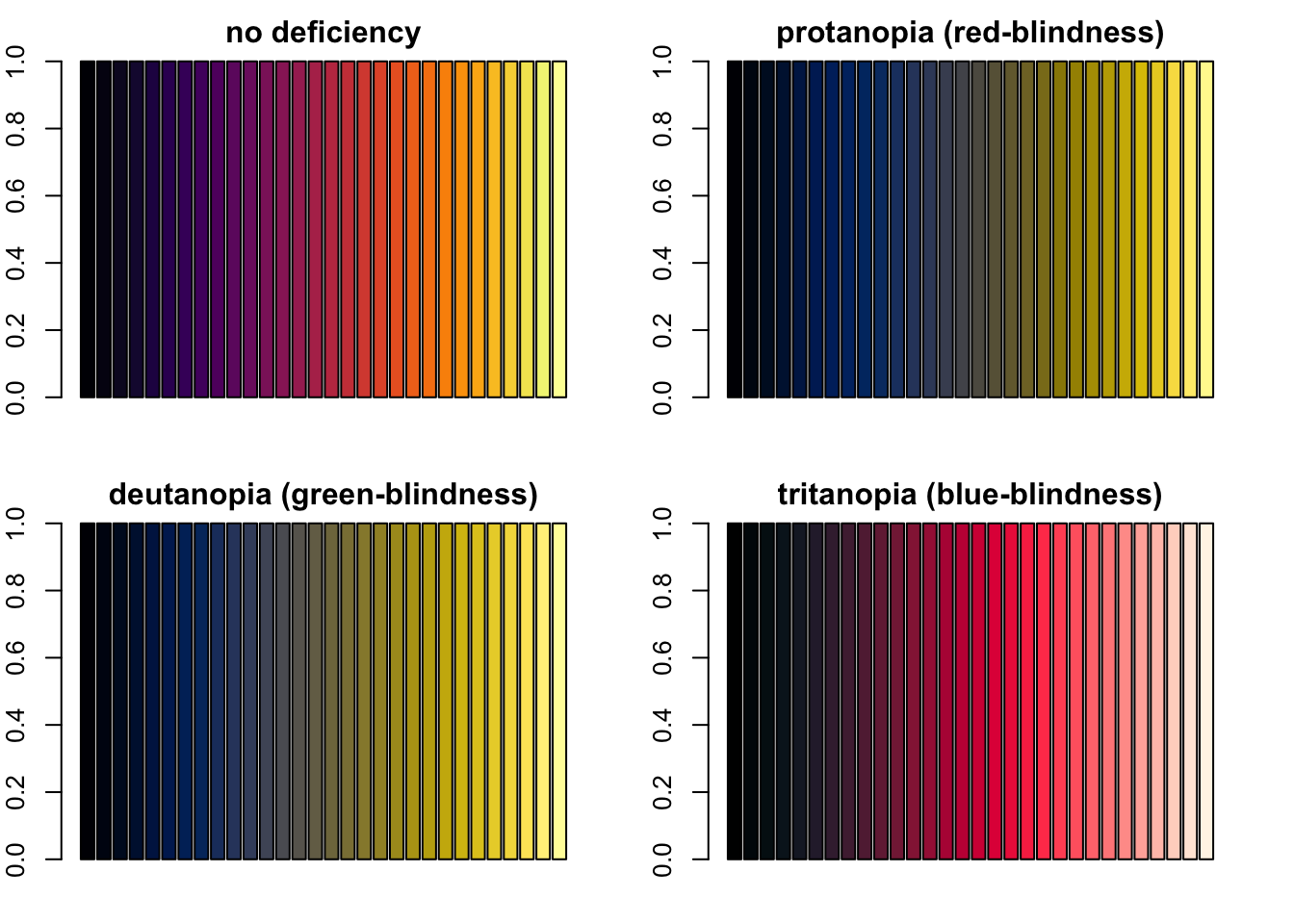
Four barplots demonstating how viridis’ inferno palette looks like with severe protanopia, deutanopia and tritanopia.
Session Info
R Markdown and the various other packages I mentioned above are changing with incredible pace, and keep getting new features. So, in all likelihood, the tips and advice above will become outdated or not necessary soon. Just in case, anyone wants to reproduce what I am talking about above here is the info about the R session (including package versions) I used to produce this page
sessionInfo()R version 4.4.1 (2024-06-14)
Platform: aarch64-apple-darwin20
Running under: macOS Sonoma 14.5
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/4.4-arm64/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/4.4-arm64/Resources/lib/libRlapack.dylib; LAPACK version 3.12.0
locale:
[1] en_GB.UTF-8/en_GB.UTF-8/en_GB.UTF-8/C/en_GB.UTF-8/en_GB.UTF-8
time zone: Europe/London
tzcode source: internal
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] viridis_0.6.5 viridisLite_0.4.2 colorspace_2.1-0 RColorBrewer_1.1-3
[5] knitr_1.46 fortunes_1.5-4
loaded via a namespace (and not attached):
[1] gtable_0.3.5 jsonlite_1.8.8 dplyr_1.1.4 compiler_4.4.1
[5] tidyselect_1.2.1 gridExtra_2.3 jquerylib_0.1.4 scales_1.3.0
[9] yaml_2.3.8 fastmap_1.2.0 ggplot2_3.5.1 R6_2.5.1
[13] generics_0.1.3 tibble_3.2.1 bookdown_0.39 munsell_0.5.1
[17] bslib_0.7.0 pillar_1.9.0 rlang_1.1.4 utf8_1.2.4
[21] cachem_1.1.0 xfun_0.43 sass_0.4.9 cli_3.6.2
[25] magrittr_2.0.3 digest_0.6.35 grid_4.4.1 lifecycle_1.0.4
[29] vctrs_0.6.5 evaluate_0.23 glue_1.7.0 blogdown_1.19
[33] fansi_1.0.6 rmarkdown_2.26 tools_4.4.1 pkgconfig_2.0.3
[37] htmltools_0.5.8.1External links and resources
colorbrewer2.org: dashboard to select palettes, with the option to limit only to colourblind-friendly ones.
RColorBrewer R package: used to generate colourblind-friendly palletes (in the form of vectors of hex colour codes). For example,
Accessible hyperlinks tutorial by NC State University can be useful.
Improve accessibility of HTML pages section in R Markdown cookbook