APTS Statistical Modelling
Beyond generalized linear models
Professor of Statistics
Department of Statistics, University of Warwick
ioannis.kosmidis@warwick.ac.uk
ikosmidis.com ikosmidis ikosmidis_
19 April 2024
Generalized linear model
Generalized linear models
Data
Response values \(y_1, \ldots, y_n\), with \(y_i \in \Re\)
Covariate vectors \(x_1, \ldots, x_n\), with \(x_i = (x_{i1}, \ldots, x_{ip})^\top \in \Re^p\)
Model
Random component
\(Y_i \mid x_i \stackrel{\text{ind}}{\sim}\mathop{\mathrm{EF}}(\mu_i,\sigma^2)\), where \(\mathop{\mathrm{EF}}(\mu_i,\sigma^2)\) is an exponential family distribution with mean \(\mu_i\) and variance \(\sigma^2 V(\mu_i) / m_i\) for known \(m_i\)
Systematic component
A linear predictor \(\eta_i = x_i^\top \beta\)
Link
\(g(\mu_i) = \eta_i\), where \(g(\cdot)\) is called the link function
Generalized linear models
Generalized linear models (GLMs) have proved effective at modelling real-world variation in a wide range of application areas.
Settings where GLMs may fail
Mean model cannot be appropriately specified due to dependence on an unobserved or unobservable covariate.
Excess variability between experimental units beyond what is implied by the mean/variance relationship of the chosen response distribution.
Assumption of independence is not appropriate.
Complex multivariate structures in the data require a more flexible model.
Overdispersion
Toxoplasmosis: ?SMPracticals::toxo
The number of people testing positive for toxoplasmosis (\(r_1, \ldots, r_n\)) out of the number of people tested (\(m_1, \ldots, m_n\)) in \(n = 34\) cities in El Salvador, along with the annual rainfall in mm in those cities (\(x_1, \ldots, x_n\)).
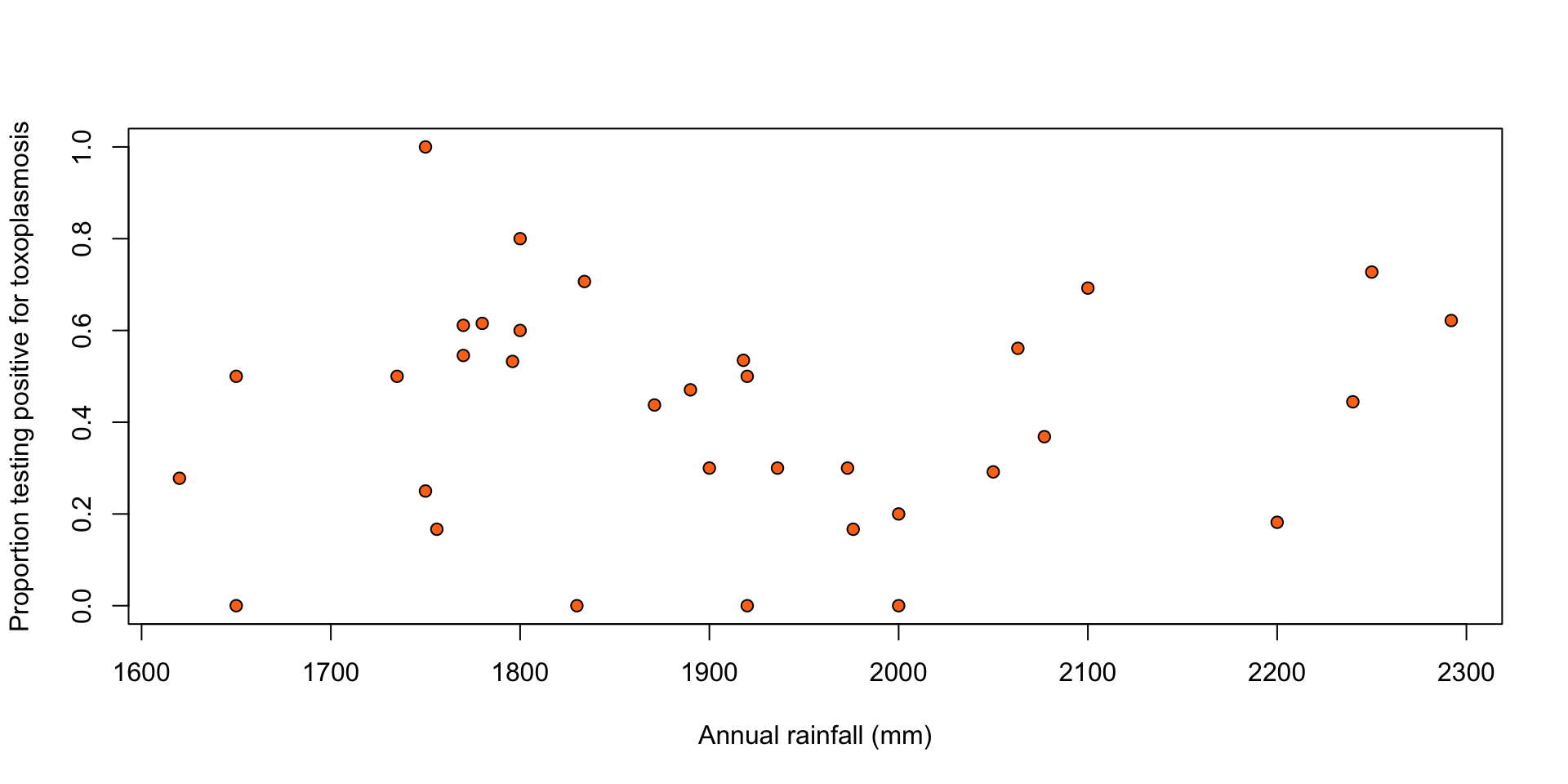
Figure 1: Proportion of people testing positive for toxoplasmosis against rainfall.
Toxoplasmosis
Logistic regression
Random component
\(Y_1, \ldots , Y_n\) are conditionally independent given the covariate values \(x_1, \ldots, x_n\), with \[ \begin{aligned} Y_i & = R_i / m_i \\ R_i \mid x_i & \sim \mathop{\mathrm{Binomial}}(m_i, \mu_i) \end{aligned} \]
Systematic component and link
\[ \log \frac{\mu_i}{1 - \mu_i} = \eta_i = \beta_1 + f(x_i) \] for some function \(f(\cdot)\) of the covariates.
Implied response variance
\[ \mathop{\mathrm{var}}(Y_i \mid x_i) = \mu_i (1 - \mu_i) / m_i \]
Toxoplasmosis
Let \(f(x_i) = 0\) (intercept only) and \(f(x_i) = \beta_1 f_1(x_i) + \beta_2 f_2(x_i) + \beta_3 f_3(x_i)\) (cubic), where \(f_1(\cdot), f_2(\cdot), f_3(\cdot)\) are orthogonal polynomials of degree 1, 2, and 3, respectively, i.e. \(\sum_{i = 1}^n f_j(x_i)^2 = 1\), and \(\sum_{i = 1}^n f_j(x_i) f_k(x_i) = 0\), for \(j \ne k\).
data("toxo", package = "SMPracticals")
mod_const <- glm(r/m ~ 1, data = toxo, weights = m,
family = "binomial")
mod_cubic <- glm(r/m ~ poly(rain, 3), data = toxo, weights = m,
family = "binomial")
plot(r/m ~ rain, data = toxo, xlab = "Annual rainfall (mm)",
ylab = "Proportion testing positive for toxoplasmosis",
bg = "#ff7518", pch = 21)
pred_cubic <- function(x)
predict(mod_cubic, newdata = list(rain = x), type = "response")
abline(h = plogis(coef(mod_const)), lty = 2)
curve(pred_cubic, add = TRUE, lty = 3)
legend("topleft", legend = c("intercept only", "cubic"), lty = c(2, 3))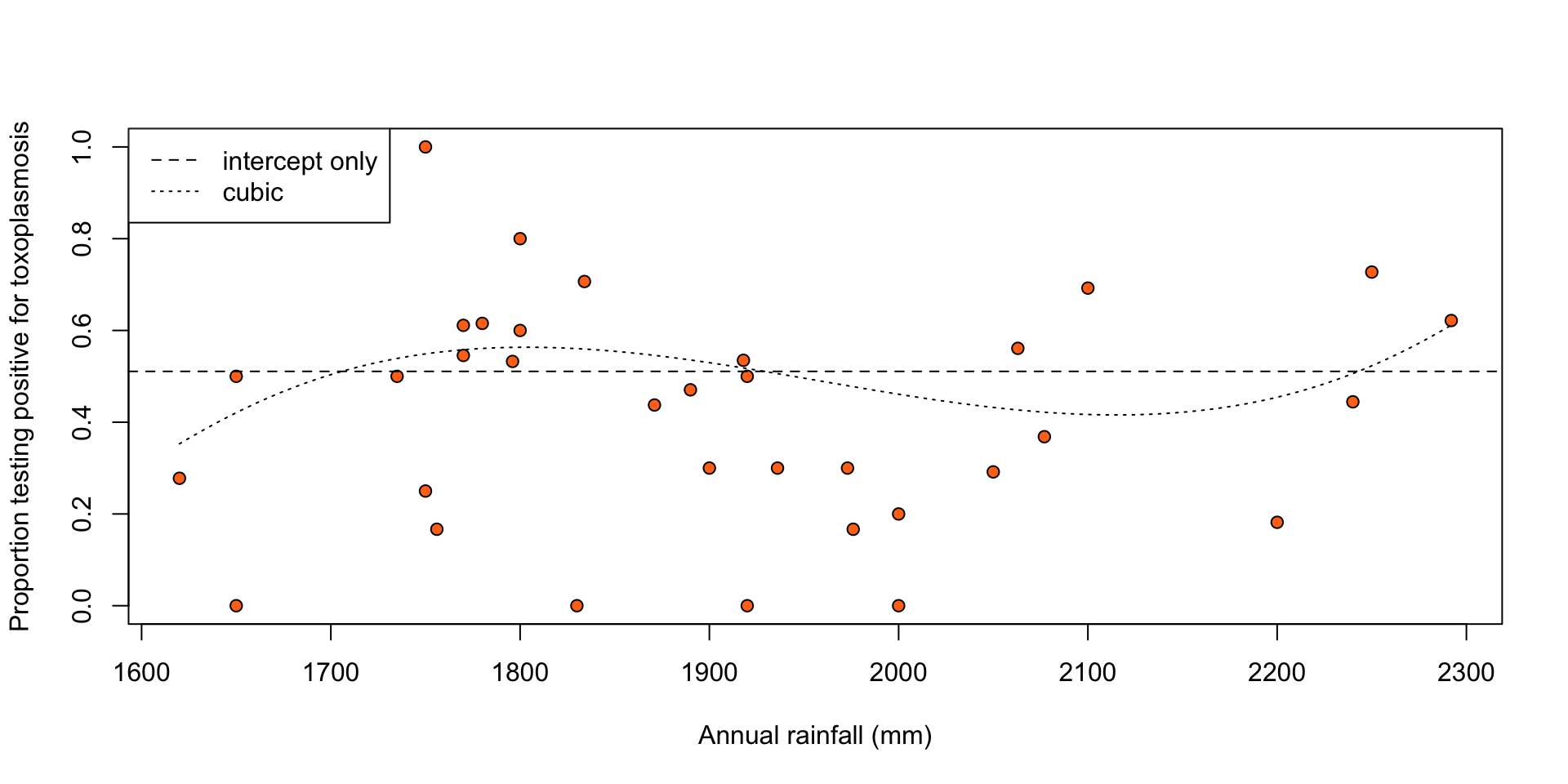
Figure 2: Proportion of people testing positive for toxoplasmosis against rainfall, with fitted proportions under an intercept-only (dashed line) and a cubic (dotted line) logistic regression model.
Toxoplasmosis
Comparison of intercept-only to cubic model
Analysis of Deviance Table
Model 1: r/m ~ 1
Model 2: r/m ~ poly(rain, 3)
Resid. Df Resid. Dev Df Deviance Pr(>Chi)
1 33 74.212
2 30 62.635 3 11.577 0.008981 **
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1But
Residual deviance for the cubic model is 62.63, which is much larger than the residual degrees of freedom (30), indicating a poor fit.
May be due to overdispersion of the observed response relative to the response variance that logistic regression prescribes.
Nelder, J. A., & Wedderburn, R. W. M. (1972). Generalized linear models. Journal of the Royal Statistical Society: Series A (General), 135(3), 370–384.
DOI: 10.2307/2344614Wedderburn, R. W. M. (1974). Quasi-Likelihood Functions, Generalized Linear Models, and the Gauss-Newton Method. Biometrika, 61(3), 439.
DOI: 10.2307/2334725
Quasi-likelihood
Model specification
Instead of specifying a full probability model, we can specify only
Mean structure
\({{\color[rgb]{0, 0.4627, 0.6313}{g}}}(\mu_i) = \eta_i = {{\color[rgb]{0, 0.4627, 0.6313}{x}}_i^\top \beta}\)
Response variance
\(\mathop{\mathrm{var}}(Y_i \mid x_i) = \sigma^2 {{\color[rgb]{0, 0.4627, 0.6313}{V}}}(\mu_i) / m_i\)
Quasi-likelihood estimation
Estimator of \(\beta\)
Solve, with respect to \(\beta\), \[ \sum_{i = 1}^n x_i {\frac{y_i-\mu_i}{\sigma^2 V(\mu_i) g'(\mu_i)/m_i}} = 0 \tag{1}\] which can be done without knowing \(\sigma^2\).
Estimator of \(\sigma^2\)
\[ \hat\sigma^2\equiv \frac{1}{n - p} \sum_{i=1}^n \frac{m_i(y_i-\hat{\mu}_i)^2}{V(\hat{\mu}_i)} \]
Quasi-likelihood
Asymptotic behaviour
If the mean and variance functions are correctly-specified, then
\[\hat\sigma (X^\top W X)^{1/2} (\hat\beta - \beta) \stackrel{d}{\rightarrow} \mathop{\mathrm{N}}(0, I_p)\]
Computation
If \(V(\mu_i)\) is the same function as in the definition of \(\mathop{\mathrm{var}}(Y_i \mid x_i)\) for an \(\mathop{\mathrm{EF}}(\mu_i, \sigma^2)\) distribution, then (1) could be solved using standard GLM routines.
Toxoplasmosis
Cubic logistic regression model
Call:
glm(formula = r/m ~ poly(rain, 3), family = "binomial", data = toxo,
weights = m)
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 0.02427 0.07693 0.315 0.752401
poly(rain, 3)1 -0.08606 0.45870 -0.188 0.851172
poly(rain, 3)2 -0.19269 0.46739 -0.412 0.680141
poly(rain, 3)3 1.37875 0.41150 3.351 0.000806 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 74.212 on 33 degrees of freedom
Residual deviance: 62.635 on 30 degrees of freedom
AIC: 161.33
Number of Fisher Scoring iterations: 3Toxoplasmosis
Quasi-likelihood for cubic model with \(\mathop{\mathrm{var}}(Y_i \mid x_i) = \sigma^2 \mu_i (1 - \mu_i) / m_i\).
Call:
glm(formula = r/m ~ poly(rain, 3), family = "quasibinomial",
data = toxo, weights = m)
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 0.02427 0.10716 0.226 0.8224
poly(rain, 3)1 -0.08606 0.63897 -0.135 0.8938
poly(rain, 3)2 -0.19269 0.65108 -0.296 0.7693
poly(rain, 3)3 1.37875 0.57321 2.405 0.0225 *
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for quasibinomial family taken to be 1.940446)
Null deviance: 74.212 on 33 degrees of freedom
Residual deviance: 62.635 on 30 degrees of freedom
AIC: NA
Number of Fisher Scoring iterations: 3Toxoplasmosis
Estimated effects
Estimates of \(\beta\) from logistic regression and quasi likelihood are the same.
Value of \(\hat\sigma^2\) is 1.94, about double than \(\sigma^2 = 1\), implied by logistic regression.
Standard errors from quasi-likelihood are inflated by a factor of \(\sqrt{1.94}\).
Comparison of intercept-only to cubic quasi-likelihood fit
Parallel to linear regression, using deviances in place of residual sums of squares, compare \[ F = \frac{(74.21 - 62.63) / (33 - 30)}{1.94} \] to quantiles of its asymptotic \(F\) distribution with 33 - 30 and 30 degrees of freedom.
Evidence in favour of an effect of rainfall on toxoplasmosis incidence is now less compelling.
Parametric models for overdispersion
To construct a full probability model that accounts for overdispersion, we need to consider the reasons for overdispersion:
Important covariates, other than rainfall, which are not observed.
Other features of the cities, possibly unobservable, all having a small individual effect on incidence, but a larger effect in combination (natural excess variability between units).
Unobserved heterogeneity
We assumed a linear predictor \(\eta_i = x_i^\top \beta\), but instead suppose that the actual predictor is \[ \eta_i^* = \eta_i + \zeta_i \] where \(\zeta_i\) may involve covariates \(z_i\) that are different than those in \(x_i\).
We can compensate for the missing term \(\zeta_i\) by assuming that it has a distribution \(F\) in the population. Then, \[ \mu_i = \mathop{\mathrm{E}}(Y_i \mid x_i, \zeta_i) = g^{-1}(\eta_i + \zeta_i) \sim G \] where \(G\) is the distribution induced by \(F\), and \[ \begin{aligned} \mathop{\mathrm{E}}(Y_i \mid x_i) & = {\mathop{\mathrm{E}}}_G(\mu_i)\\ \mathop{\mathrm{var}}(Y_i \mid x_i) & = {\mathop{\mathrm{E}}}_G( \mathop{\mathrm{var}}(Y_i \mid x_i, \zeta_i) ) + {\mathop{\mathrm{var}}}_G(\mu_i) \end{aligned} \] If \(Y_i \mid x_i, \zeta_i \sim \mathop{\mathrm{EF}}(\mu_i, \sigma^2)\), then \(\mathop{\mathrm{var}}(Y_i \mid x_i) = \sigma^2 {\mathop{\mathrm{E}}}_G(V(\mu_i)) / m_i + {\mathop{\mathrm{var}}}_G(\mu_i)\).
Direct models
One approach is to model the \(Y_i\) directly, by specifying an appropriate form for \(G\).
Direct models
Beta-binomial model
\[ \begin{aligned} Y_i & = R_i / m_i \\ R_i \mid \mu_i & \stackrel{\text{ind}}{\sim}\mathop{\mathrm{Binomial}}(m_i, \mu_i) \\ \mu_i \mid x_i & \stackrel{\text{ind}}{\sim}\mathop{\mathrm{Beta}}(k \mu^*_i, k (1 - \mu^*_i)) \\ \log \{ \mu_i^* / (1 - \mu_i^*) \} & = \eta_i \end{aligned} \] \[\LARGE\Downarrow\] \[ \mathop{\mathrm{E}}(Y_i \mid x_i) = \mu^*_i \quad \text{and} \quad \mathop{\mathrm{var}}(Y_i \mid x_i) = \frac{\mu^*_i (1-\mu^*_i)}{m_i} \left(1 + {{\color[rgb]{0.70,0.1,0.12}{\frac{m_i - 1}{k + 1}}}} \right) \,, \]
Negative binomial model
A popular model for overdispersion in count responses assumes a Poisson distribution with a mean with a gamma distribution, leading to a negative binomial marginal response distribution.
Direct models
Estimation
Direct models for overdispersion can, in principled, be estimated with standard estimation methods, such as maximum likelihood. For example, the beta-binomial model has likelihood proportional to \[ \prod_{i = 1}^n \frac{\Gamma(k\mu_i^* + m_i y_i) \Gamma(k (1 - \mu_i^*) + m_i (1 - y_i)) \Gamma(k)}{ \Gamma(k\mu_i^*)\Gamma(k ( 1 - a\mu_i^*))\Gamma(k + m_i)} \,. \]
Disadvantages
Maximization of the likelihood can be numerically difficult when the parameters of the mean distribution are free.
Direct models tend to have limited flexibility on the direction of overdispersion.
Random effects models for overdispersion
Linear predictor with random effects \(b_i\)
\[ \eta_i = x_i^\top\beta + b_i \] where \(b_1, \ldots, b_n\) represent the extra, unexplained by the covariates, variability between units.
Distribution of random effects
Typically, \(b_1, \ldots, b_n\) are assumed independent with \(b_i \sim {\mathop{\mathrm{N}}}(0,\sigma^2_b)\), for some unknown \(\sigma^2_b\).
We set \(\mathop{\mathrm{E}}(b_i) = 0\), because an unknown mean for \(b_i\) would be unidentifiable in the presence of the intercept parameter in \(\eta_i\).
Random effects: likelihood
Denote \(f(y_i \mid x_i, b_i \,;\,\theta)\) the density or probability mass function of a GLM with linear predictor \(\eta_i = x_i^\top \beta + b_i\), and \(f(b_i \mid \sigma^2_b)\) the density of a \(\mathop{\mathrm{N}}(0, \sigma^2_u)\) distribution.
The likelihood of the random effects model is \[ \begin{aligned} f(y\mid X \,;\,\theta, \sigma^2_b)&=\int f(y \mid X, b \,;\,\theta, \sigma^2_u)f(b \mid X \,;\,\theta, \sigma^2_b)db\\ &=\int f(y \mid X, b \,;\,\theta)f(b \,;\,\sigma^2_b) db \\ &=\int \prod_{i=1}^n f(y_i \mid x_i, b_i \,;\,\theta) f(b_i \,;\,\sigma^2_b)db_i \end{aligned} \]
Depending on what \(f(y_i \mid x_i, b_i \,;\,\theta)\) is, no further simplification may be possible, and computation needs careful consideration.
Dependence
Toxoplasmosis
Data
The number of people testing positive \((r_i)\) out of those tested \((m_i)\) in the \(i\)th city, and the city’s annual rainfall in mm \((x_i)\) \((i = 1, \ldots, n)\).
Model
\(Y_1, \ldots , Y_n\) are conditionally independent given \(x_1, \ldots, x_n\), with \[ \begin{aligned} Y_i & = R_i / m_i \\ R_i \mid x_i & \sim \mathop{\mathrm{Binomial}}(m_i, \mu_i) \end{aligned} \]
\[ \log \frac{\mu_i}{1 - \mu_i} = \beta_1 + f(x_i) \]
\[\LARGE\equiv\]
Data
Whether the \(j\)th individual in the \(i\)th city is testing positive or not \((Y_{ij})\), and the city’s annual rainfall in mm \((x_i)\) \((i = 1, \ldots, n; j = 1, \ldots, m_i)\).
Model
\(Y_{11}, \ldots , Y_{nm_n}\) are conditionally independent given \(x_1, \ldots, x_n\), with \[ \begin{aligned} Y_{ij} \mid x_i & \sim \mathop{\mathrm{Bernoulli}}(\mu_{ij}) \end{aligned} \]
\[ \log \frac{\mu_{ij}}{1 - \mu_{ij}} = \beta_1 + f(x_i) \]
Toxoplasmosis: Overdispersion through dependence
Suppose that the only assumptions we are confident in making are:
\(R_1, \ldots, R_n\) are conditionally independent given \(x_1, \ldots, x_n\)
\(\mathop{\mathrm{cov}}(Y_{ij}, Y_{ik} \mid x_i) \ne 0\), \(j \ne k\)
Then,
\[ \begin{aligned} \mathop{\mathrm{var}}(Y_i \mid x_i) & = {\color[rgb]{1,1,1}{\frac{1}{m_i^2}\left\{\sum_{j=1}^{m_i} \mathop{\mathrm{var}}(Y_{ij} \mid x_i) + \sum_{j \ne k} \mathop{\mathrm{cov}}(Y_{ij}, Y_{ik} \mid x_i)\right\}}} \\ & {\color[rgb]{1,1,1}{= \frac{\mu_i (1 - \mu_i)}{m_i} + \frac{1}{m_i^2} \sum_{j \ne k} \mathop{\mathrm{cov}}(Y_{ij}, Y_{ik} \mid x_i)}} \\ \end{aligned} \]
Positive correlation between individuals in the same city induces overdispersion in the number of positive cases.
Dependence: Unobserved heterogeneity
There is a number of plausible reasons why there is dependence between responses corresponding to units within a given cluster (in the toxoplasmosis example, clusters are cities). One compelling reason is unobserved heterogeneity.
For the toxoplasmosis status of individuals within a city it may hold \[ Y_{ij} \perp\!\!\!\perp Y_{ik} \mid x_i, \zeta_i \quad (j \ne k) \] where \(\zeta_i\) is the unobserved part of a true linear predictor. However, \[ Y_{ij} \perp\!\!\!\perp\!\!\!\!\!\!/\;\;Y_{ik} \mid x_i \quad (j \ne k) \]
Conditional (given \(\zeta_i\)) independence between units in a common cluster \(i\) becomes marginal dependence, when marginalized over the population distribution \(F\) of \(\zeta_i\).
Random effects and dependence
For the toxoplasmosis status of individuals within a city, we can account for intra-cluster dependence, through the model \[ \begin{aligned} Y_{ij} \mid x_{i}, b_i & \stackrel{\text{ind}}{\sim}\mathop{\mathrm{Bernoulli}}(\mu_{ij}) \,, \\ \log \frac{\mu_{ij}}{1-\mu_{ij}} & = \beta_1 + f(x_i) + b_i\,, \\ b_i & \stackrel{\text{ind}}{\sim}{\mathop{\mathrm{N}}}(0, \sigma^2_b)\,, \end{aligned} \] which, for \(\sigma^2_b > 0\), implies \[ Y_{ij} \perp\!\!\!\perp\!\!\!\!\!\!/\;\;Y_{ik} \mid x_{i} \]
Marginal models
Model specification
Instead of specifying a full probability model, we can specify a marginal model, through
Mean structure
\(\displaystyle \begin{aligned} \mu_{ij} & = \mathop{\mathrm{E}}(Y_{ij} \mid \eta_{ij}) \\ g(\mu_{ij}) & = \eta_{ij} = x_{ij}^T\beta \end{aligned}\)
Response variance
\(\displaystyle \mathop{\mathrm{var}}(Y_{ij} \mid x_{ij})= \sigma^2 V(\mu_{ij}) / m_{ij}\)
Correlation between responses
\(\displaystyle \mathop{\mathrm{corr}}(Y_{ij}, Y_{ik} \mid x_{ij}, x_{ik}) = \rho(\mu_{i}, \psi)\)
with \(\mu_i = (\mu_{i1}, \ldots, \mu_{in_i})^\top\), where \(n_i\) is the number of observations in the \(i\)th cluster.
Marginal models versus random effects
The parameters \(\beta\) in a marginal model have a different interpretation from those in a random effects model because \[ \mathop{\mathrm{E}}(Y_{ij} \mid x_{ij}) = \mathop{\mathrm{E}}(g^{-1}[x_{ij}^\top\beta + b_i]) \ne g^{-1}(x_{ij}^\top\beta) \quad \text{if $g$ is not linear} \]
A random effects model describes the mean response at the subject level (subject specific).
a marginal model describes the mean response across the population (population averaged).
Generalized estimating equations (GEEs)
Estimator of \(\beta\)
Solve, with respect to \(\beta\), \[ \sum_i \left(\frac{\partial\mu_i}{\partial\beta}\right)^T\mathop{\mathrm{var}}(Y_i)^{-1}(Y_i-\mu_i) = 0 \] where \(Y_i = (Y_{i1}, \ldots, Y_{in_i})^\top\).
Notes
There are several consistent estimators for the covariance of GEE estimators.
The GEE approach is generally robust to misspecification of the correlation structure.
Clustered data
A few examples where data are collected in clusters include:
Studies in biometry where repeated measurements are made on experimental units. Such studies can effectively mitigate the effect of between-unit variability on important inferences.
Agricultural field trials, or similar studies, for example in engineering, where experimental units are arranged within blocks.
Sample surveys where collecting data within clusters or small areas can save costs.
Data collected in space or time, where spatial or serial dependence is induced by the arrangement of units of observation.
Rat growth: ?SMPracticals::rat.growth
The weekly weights of 30 young rats.
data("rat.growth", package = "SMPracticals")
plot(y ~ week, data = rat.growth, xlab = "Week", ylab = "Weight",
type = "n")
for (i in 1:30) {
dat_i <- subset(rat.growth, rat == i)
lines(y ~ week, data = dat_i, col = "grey")
}
points(y ~ week, data = rat.growth, xlab = "Week", ylab = "Weight",
bg = "#ff7518", pch = 21, col = "grey")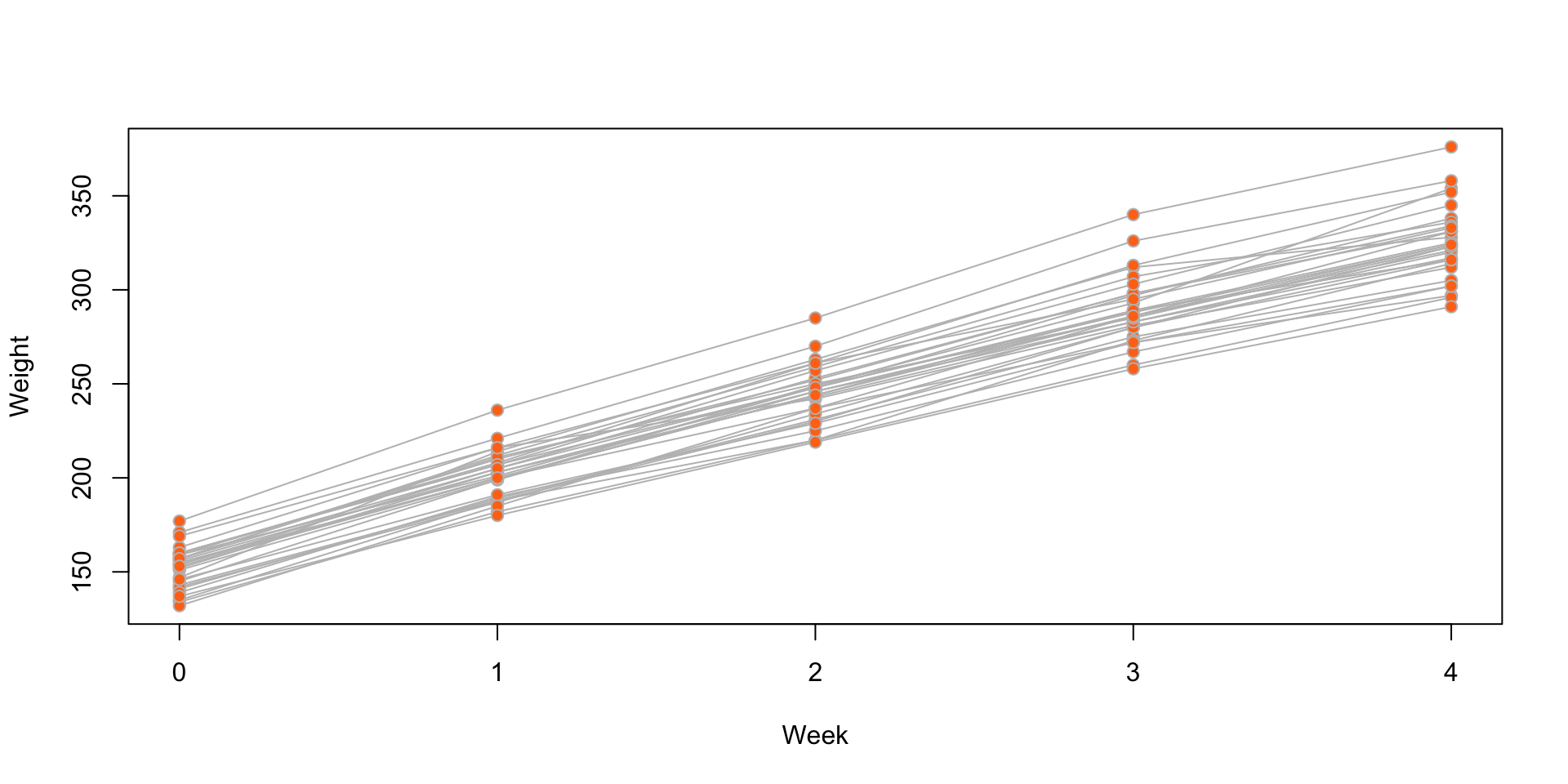
Figure 3: Individual rat weight by week, for the rat growth data.
Rat growth: Simple linear regression model
Let \(y_ij\) be the weight of rat \(i\) at week \(x_{ij}\), and consider the simple linear regression model \[ Y_{ij} = \beta_1 + \beta_2 x_{ij} + \epsilon_{ij} \]
Call:
lm(formula = y ~ week, data = rat.growth)
Residuals:
Min 1Q Median 3Q Max
-38.12 -11.25 0.28 7.68 54.15
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 156.053 2.246 69.47 <2e-16 ***
week 43.267 0.917 47.18 <2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 15.88 on 148 degrees of freedom
Multiple R-squared: 0.9377, Adjusted R-squared: 0.9372
F-statistic: 2226 on 1 and 148 DF, p-value: < 2.2e-16Rat growth: Residual analysis
res <- residuals(rat_lm, type = "pearson")
ord <- order(ave(res, rat.growth$rat))
rats <- rat.growth$rat[ord]
rats <- factor(rats, levels = unique(rats), ordered = TRUE)
plot(res[ord] ~ rats,
xlab = "Rat (ordered by mean residual)",
ylab = "Pearson residual",
col = "#ff7518", pch = 21)
abline(h = 0, lty = 2)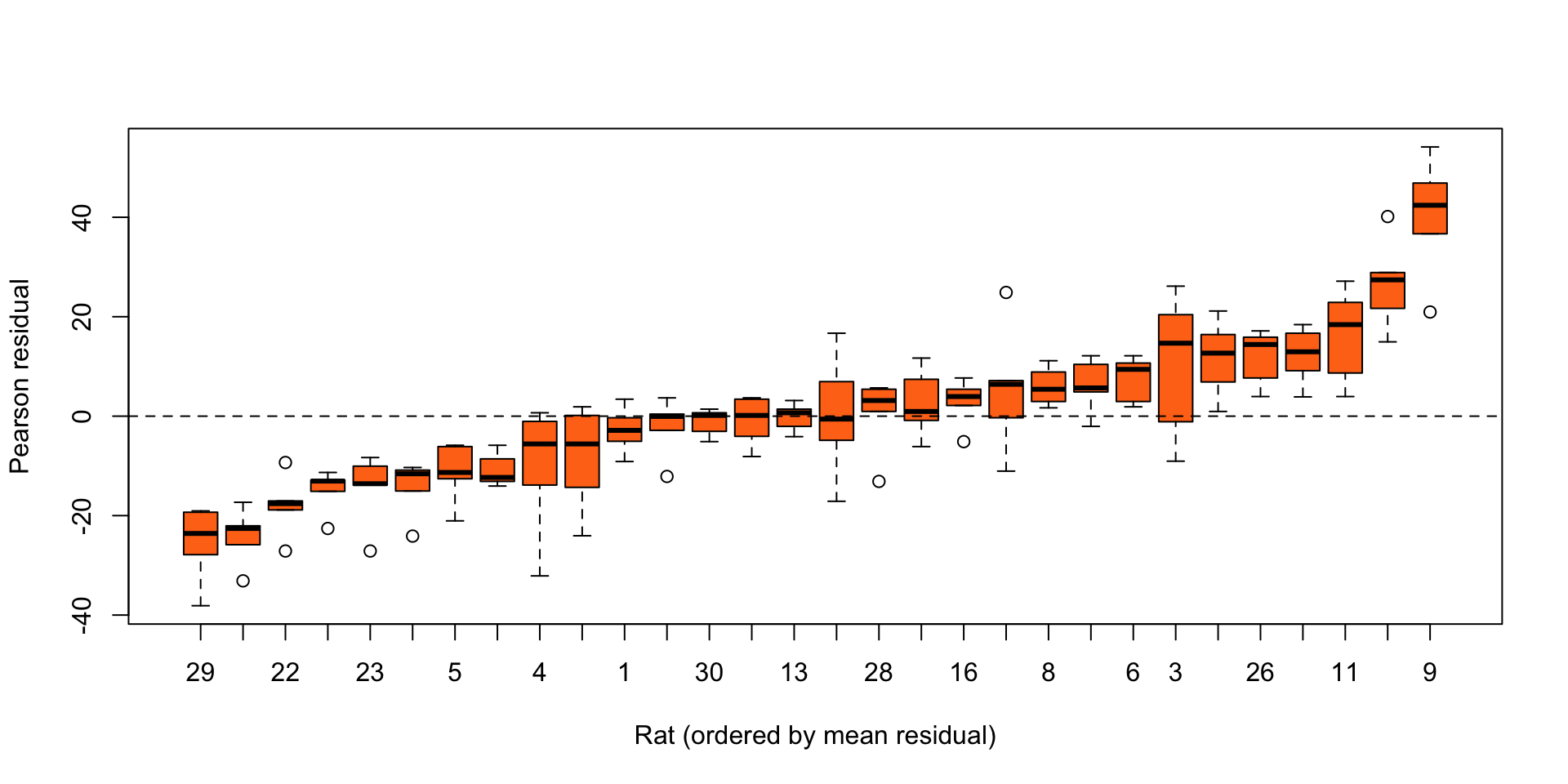
Figure 4: Boxplots of residuals from a simple linear regression, for each rat in the rat growth data.
Clear evidence of unexplained differences between rats.
Linear mixed models
Linear mixed models
Data
Response values \(y = (y_1, \ldots , y_n)^\top\)
Covariate matrix \(X^*\) of dimension \(n \times p^*\)
Linear mixed model (LMM)
\[ \begin{aligned} Y \mid X, Z, b & \sim {\mathop{\mathrm{N}}}(\mu, \Sigma) \,, \quad \mu = X\beta + Z b \\ b & \sim {\mathop{\mathrm{N}}}(0, \Sigma_b) \end{aligned} \tag{2}\] where \(X\) and \(Z\) are \(n \times p\) and \(n \times q\) matrices, respectively, formed by columns of \(X^*\).
The unknown parameters are \(\beta\), \(\Sigma_b\), and \(\Sigma\) (we typically assume that \(\Sigma = \sigma^2 I_n\)).
An LMM for clustered data
\[ \begin{aligned} Y_{ij} \mid x_{ij}, z_{ij}, b_i & \stackrel{\text{ind}}{\sim}{\mathop{\mathrm{N}}}(\mu_{ij},\sigma^2) \\ \mu_{ij} & = x_{ij}^T \beta + z_{ij}^\top b_i \\ b_i & \stackrel{\text{ind}}{\sim}{\mathop{\mathrm{N}}}(0,\Sigma^*_b) \end{aligned} \] where \(x_{ij}\) contains the covariates for observation \(j\) of the \(i\)th cluster, and \(z_{ij}\) the covariates that exhibit extra between-cluster variation in their relationship with the response.
\(z_{ij}\) may or may not be a sub-vector of \(x_{ij}\).
Random intercept model
\(z_{ij} = 1\) results in \(\mu_{ij} = x_{ij}^\top \beta + b_i\)
Random intercept and slope model with a single covariate
\[ Y_{ij} = \beta_1 + b_{1i} + (\beta_2 + b_{2i}) x_{ij} + \epsilon_{ij} \quad (b_{1i},b_{2i})^\top \stackrel{\text{ind}}{\sim}{\mathop{\mathrm{N}}}(0, \Sigma^*_b) \]
Fits in the general LMM definition in (2) with \(\Sigma = \sigma^2 I_n\), and
\[ Y = \begin{bmatrix} Y_1 \\ \vdots \\ Y_k \end{bmatrix}\,, \quad Y_i = \begin{bmatrix} Y_{i1} \\ \vdots \\ Y_{in_i} \end{bmatrix}\,, \quad X = \begin{bmatrix} X_1 \\ \cdots \\ X_k \end{bmatrix}\,, \quad Z = \begin{bmatrix} X_1 & 0 & \cdots & 0 \\ 0 & X_2 & \cdots & 0 \\ \vdots & \vdots & \ddots & \vdots \\ 0 & 0 & \cdots & X_k \end{bmatrix}\,, \quad X_i = \begin{bmatrix} 1 & x_{i1} \\ \vdots & \vdots \\ 1 & x_{in_i} \end{bmatrix} \] \[ \beta = \begin{bmatrix} \beta_1 \\ \beta_2 \end{bmatrix}\,, \quad b = \begin{bmatrix} b_{11} \\ b_{21} \\ \vdots \\ b_{1k} \\ b_{2k} \end{bmatrix}\,, \quad \Sigma_b = \begin{bmatrix} \Sigma^*_b & 0 & 0 \\ 0 & \ddots & 0 \\ 0 & 0 & \Sigma^*_b \end{bmatrix} \] where \(\Sigma^*_b\) is a \(2\times 2\) positive definite matrix, and \(0\) is a matrix of zeros of appropriate dimension.
Variance components
Mixed model
\[ X\underbrace{\beta}_{\text{fixed effects}} + Z \underbrace{b}_{\text{random effects}} \]
Marginal distribution
Under the LMM in (2), the marginal distribution of \(Y\) is \[ Y \mid X, Z \sim {\mathop{\mathrm{N}}}(X \beta,\Sigma + Z\Sigma_b Z^\top ) \tag{3}\]
Hence, \(\mathop{\mathrm{var}}(Y \mid X, Z)\) is comprised of two variance components.
Hierarchical or multilevel models
LMMs for clustered data are also known as hierarchical or multilevel models.
This reflects the two-stage structure in their definition; a conditional model for the responses given covariates and the random effects, followed by a marginal model for the random effects.
The hierarchy can have further levels, indicating clusters nested within clusters, e.g. patients within wards within hospitals, pupils within classes within schools within cities, etc.
Random effects or cluster-specific fixed effects
Cluster-specific random effects
\(Y_{ij} = \beta_1 + b_{1i} + (\beta_2 + b_{2i})x_{ij} +\epsilon_{ij}\)
Require additional assumptions about the distribution of the random effects.
Allow inferences to be extended to a wider population.
Random effects are generally identifiable and can be estimated.
Random effects allow borrowing strength across clusters by shrinking fixed effects towards a common mean.
Cluster-specific fixed effects
\(Y_{ij} = \beta_{1i} + \beta_{2i} x_{ij} + \epsilon_{ij}\)
No need for additional distributional assumptions.
Inferences can only be made about those clusters present in the observed data.
Fixed effects may not be identifiable (e.g. one observation per cluster).
The asymptotic behaviour of standard estimators may not be what is expected by standard theory.
Maximum likelihood estimation
Likelihood about \(\beta, \Sigma, \Sigma_b\)
\[ f(y \mid X, Z \,;\,\beta, \Sigma, \Sigma_b) \propto |V|^{-1/2} \exp\left(-\frac{1}{2} (y-X\beta)^\top V^{-1} (y-X\beta)\right) \tag{4}\] where \(V = \Sigma + Z\Sigma_b Z^\top\).
(4) can be directly maximized using numerical optimization techniques, exploiting the fact that for fixed \(V\), the maximum likelihood estimator of \(\beta\) is \[ \hat\beta_V = (X^\top V^{-1} X)^{-1}X^\top V^{-1} Y \]
Estimation of variance parameters
The maximum likelihood estimators for the variance parameters of LMMs can have considerable downward bias.
REML
REstricted or REsidual Maximum Likelihood (REML) proceeds by estimating the variance parameters \(\Sigma\) and \(\Sigma_b\) using a marginal likelihood based on the residuals from a least squares fit of the model \(E(Y \mid X) = X \beta\).
Marginal likelihood
\[ \frac{|X^\top X|^{1/2}}{|V|^{1/2} |X^\top V^{-1} X|^{1/2}} \exp\left(-\frac{1}{2} (y-X\hat\beta_V)^\top V^{-1} (y-X\hat\beta_V)\right) \tag{5}\]
REML derivation: Residuals
\[ \begin{aligned} R = (I_n - H) Y & = {\color[rgb]{1,1,1}{(I_n - H) (X\beta + A)}} \\ & {\color[rgb]{1,1,1}{= X\beta - \underbrace{X (X^\top X)^{-1} X^\top}_{\text{hat matrix}} X \beta + (I_n - H) A}} \\ & = (I_n - H) A\,, \quad \text{where } A \mid Z \sim {\mathop{\mathrm{N}}}(0, V) \end{aligned} \]
\(R\) does not depend of \(\beta\), but its distribution is degenerate because \((I_n - H)\) has rank \(n - p\).
REML derivation: Linearly independent residuals
By idempotence, \((I_n - H)\) has eigenvalues \({\color[rgb]{1,1,1}{0}}\) or \({\color[rgb]{1,1,1}{1}}\). Because it is also symmetric, its spectral decomposition gives an \(n \times (n - p)\) matrix \(B\), with \(B^\top B = I_{n - p}\), and \[ (I_n - H) = {\color[rgb]{1,1,1}{\begin{bmatrix} B & B_* \end{bmatrix} \begin{bmatrix} I_{n - p} & 0 \\ 0 & 0 \end{bmatrix} \begin{bmatrix} B^\top \\ B_*^\top \end{bmatrix}}} = B B^\top \]
So, \(U = B^\top Y\) is a vector a linearly independent residuals, and \[ \begin{aligned} U & = B^\top Y \\ & {\color[rgb]{1,1,1}{= B^\top (X \beta + A)}} \\ & {\color[rgb]{1,1,1}{= B^\top B B^\top X \beta + B^\top A}} \\ & {\color[rgb]{1,1,1}{= B^\top (I_n - H) X \beta + B^\top A}} \\ & = B^\top A \end{aligned} \]
So, \(U \mid X, Z \sim \mathop{\mathrm{N}}(0, B^\top V B)\), which does not depend on \(\beta\).
But, it seems as if dependence on \(\beta\) has been traded for dependence on \(B\), which is not uniquely defined…
REML derivation: Independence of \(U\) and \(\hat\beta_V - \beta\)
Distribution of least squares estimator of \(\beta\) for known \(V\)
\[ \begin{aligned} \hat\beta_V & = (X^\top V^{-1} X)^{-1}X^\top V^{-1} Y \\ & {\color[rgb]{1,1,1}{= (X^\top V^{-1} X)^{-1}X^\top V^{-1} (X\beta + A)}} \\ & = \beta + (X^\top V^{-1} X)^{-1}X^\top V^{-1} A \end{aligned} \]
So, \(\hat\beta_V - \beta \mid X, Z \sim {\mathop{\mathrm{N}}}(0, (X^\top V^{-1} X)^{-1})\)
Independence of \(U\) and \(\hat\beta_V - \beta\)
\[ \begin{aligned} \mathop{\mathrm{E}}(U (\hat\beta_V - \beta)^\top \mid X, Z) & = {\color[rgb]{1,1,1}{B^\top \mathop{\mathrm{E}}(A A^\top \mid Z) V^{-1} X (X^\top V^{-1}X)^{-1}}} \\ & = 0 \end{aligned} \]
Since, both \(U\) and \(\hat\beta - \beta\) are normal, they are independent!
REML derivation: Linear transformation of the response
Let \(Q = \begin{bmatrix} B^\top \\ G^\top \end{bmatrix}\), where \(G^\top = (X^\top V^{-1} X)^{-1}X^\top V^{-1}\). Then \[ Q Y = \begin{bmatrix} U \\ \hat\beta_V \end{bmatrix} \]
Temporarily suppressing the conditioning on \(X\) and \(Z\) in the notation, we can write \[ \begin{aligned} f(u \,;\,\Sigma, \Sigma_b) f(\hat\beta_V \,;\,\beta, \Sigma, \Sigma_b) & = {\color[rgb]{1,1,1}{f(u, \hat\beta_V \,;\,\beta, \Sigma, \Sigma_b)}} \\ & {\color[rgb]{1,1,1}{= f(Q y \,;\,\beta, \Sigma, \Sigma_b)}} \\ & = f(y \,;\,\beta, \Sigma, \Sigma_b) \underbrace{| Q^\top |^{-1}}_{\text{Jacobian of } Qy} \end{aligned} \]
REML derivation: Jacobian of \(Qy\)
\[ \begin{aligned} | Q^\top | & = \left|\begin{bmatrix} B & G \end{bmatrix}\right| \\ & {\color[rgb]{1,1,1}{= \left| \begin{bmatrix} B^\top \\ G^\top \end{bmatrix} \begin{bmatrix} B & G \end{bmatrix}\right|^{1/2}}} \\ & {\color[rgb]{1,1,1}{= \left| \begin{bmatrix} B^\top B & B^\top G \\ G^\top B & G^\top G \end{bmatrix} \right|^{1/2}}} \\ & {\color[rgb]{1,1,1}{= |B^\top B|^{1/2} |G^\top G - G^\top B (B^\top B)^{-1} B^\top G|^{1/2}}} \\ & {\color[rgb]{1,1,1}{= |G^\top G - G^\top (I_n - H) G|^{1/2}}} \\ & {\color[rgb]{1,1,1}{= |(X^\top V^{-1} X)^{-1}X^\top V^{-1} X (X^\top X)^{-1} X^\top V^{-1} X (X^\top V^{-1} X)^{-1} |^{1/2}}} \\ & = |X^\top X|^{-1/2} \end{aligned} \]
REML derivation: Marginal likelihood
\[ \begin{aligned} f(u \,;\,\Sigma, \Sigma_b) f(\hat\beta_V \,;\,\beta, \Sigma, \Sigma_b) & = f(y \,;\,\beta, \Sigma, \Sigma_b) | Q^\top |^{-1} \implies \\ f(u \,;\,\Sigma, \Sigma_b) & = \frac{f(y \,;\,\beta, \Sigma, \Sigma_b)}{f(\hat\beta_V \,;\,\beta, \Sigma, \Sigma_b)} | X^\top X |^{1/2} \implies \end{aligned} \] \[ f(u \,;\,\Sigma, \Sigma_b) \propto \frac{|X^\top X|^{1/2}}{|V|^{1/2} |X^\top V^{-1} X|^{1/2}} \exp\left(-\frac{1}{2} (y-X\hat\beta_V)^\top V^{-1} (y-X\hat\beta_V)\right) \]
Note on model choice
The maximized marginal likelihood cannot be used to compare different fixed effects specifications, due to the dependence of \(U\) on \(X\).
Estimating random effects
Best Linear Unbiased Predictor for \(b\)
A natural predictor \(\tilde b\) of the random effect vector \(b\) is obtained by minimizing the mean squared prediction error \(\mathop{\mathrm{E}}((\tilde b - b)^\top (\tilde b - b) \mid X, Z)\) where the expectation is over both \(b\) and \(Y\). Then, \[ \tilde b = \mathop{\mathrm{E}}(b\mid Y, X, Z) = (Z^\top \Sigma^{-1} Z + \Sigma_b^{-1})^{-1} Z^\top \Sigma^{-1}(Y-X\beta) \] which is the Best Linear Unbiased Predictor (BLUP) for \(b\).
Variance of \(b\)
\[ \mathop{\mathrm{var}}(b \mid Y, X, Z) = (Z^\top \Sigma^{-1} Z + \Sigma_b^{-1})^{-1} \]
\(\mathop{\mathrm{E}}(b\mid Y, X, Z)\) and \(\mathop{\mathrm{var}}(b\mid Y, X, Z)\) are estimated by plugging in \(\hat\beta\), \(\hat\Sigma\), and \(\hat\Sigma_b\).
\(\tilde{b}\) typically shrink towards 0 relative to the corresponding fixed effects estimates.
Estimating random effects
Any component \(b_k\) of \(b\) with no relevant data (e.g. a cluster effect for an as yet unobserved cluster) corresponds to a null column of \(Z\).
In that case, \(\tilde b_k = 0\) and \(\mathop{\mathrm{var}}(b_k \mid Y, X, Z) = [\Sigma_b]_{kk}\), which can be estimated in the common case that \(b_k\) shares a variance with other random effects.
Rat growth: Random cluster-specific intercept and slope
\[ Y_{ij} = \beta_1 + b_{1i} + (\beta_2 + b_{2i}) x_{ij} +\epsilon_{ij}\,,\quad (b_{1i},b_{2i})^\top \stackrel{\text{ind}}{\sim}{\mathop{\mathrm{N}}}(0,\Sigma_b)\,,\quad \epsilon_{ij} \stackrel{\text{ind}}{\sim}{\mathop{\mathrm{N}}}(0,\sigma^2) \]
Linear mixed model fit by REML ['lmerMod']
Formula: y ~ week + (week | rat)
Data: rat.growth
REML criterion at convergence: 1084.58
Random effects:
Groups Name Std.Dev. Corr
rat (Intercept) 10.933
week 3.535 0.18
Residual 5.817
Number of obs: 150, groups: rat, 30
Fixed Effects:
(Intercept) week
156.05 43.27 Rat growth: Random cluster-specific intercept
\[ Y_{ij} = \beta_1 + b_{1i} + \beta_2 x_{ij} + \epsilon_{ij}\,,\quad (b_{1i},b_{2i})^\top \stackrel{\text{ind}}{\sim}{\mathop{\mathrm{N}}}(0,\Sigma_b)\,,\quad \epsilon_{ij} \stackrel{\text{ind}}{\sim}{\mathop{\mathrm{N}}}(0,\sigma^2) \]
Linear mixed model fit by REML ['lmerMod']
Formula: y ~ week + (1 | rat)
Data: rat.growth
REML criterion at convergence: 1127.169
Random effects:
Groups Name Std.Dev.
rat (Intercept) 13.851
Residual 8.018
Number of obs: 150, groups: rat, 30
Fixed Effects:
(Intercept) week
156.05 43.27 Rat growth: Model comparison
The two models can be compared using AIC or BIC, but, for that, we need to refit the models with maximum likelihood rather than REML.
Data: rat.growth
Models:
rat_ri_ML: y ~ week + (1 | rat)
rat_rs_ML: y ~ week + (week | rat)
npar AIC BIC logLik deviance Chisq Df Pr(>Chisq)
rat_ri_ML 4 1139.2 1151.2 -565.60 1131.2
rat_rs_ML 6 1101.1 1119.2 -544.56 1089.1 42.079 2 7.288e-10 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Evidence is in favour of the model with random slopes.
Rat growth: fixed vs random effects
ranef_est <- coef(rat_rs)$rat
rat_lm3 <- lm(y ~ rat * week, data = rat.growth)
rats <- factor(1:30, levels = 1:30)
pred_rat_0 <- predict(rat_lm3,
newdata = data.frame(rat = rats, week = 0))
pred_rat_1 <- predict(rat_lm3,
newdata = data.frame(rat = rats, week = 1))
fixef_est <- data.frame("(Intercept)" = pred_rat_0,
"week" = pred_rat_1 - pred_rat_0)
intercept_range <- range(c(fixef_est[,1], ranef_est[,1]))
slope_range <- range(c(fixef_est[,2], ranef_est[,2]))
par(mfrow = c(1, 2))
plot(fixef_est[,1], ranef_est[,1],
xlab = "Intercept estimate (fixed effects)",
ylab = "Intercept estimate (random effects)",
xlim = intercept_range, ylim = intercept_range,
bg = "#ff7518", pch = 21)
abline(lm(ranef_est[,1] ~ fixef_est[,1]), col = "grey")
abline(a = 0, b = 1, lty = 2)
plot(fixef_est[,2], ranef_est[,2],
xlab = "Slope estimate (fixed effects)",
ylab = "Slope estimate (random effects)",
xlim = slope_range,
ylim = slope_range,
bg = "#ff7518", pch = 21)
abline(lm(ranef_est[,2] ~ fixef_est[,2]), col = "grey")
abline(a = 0, b = 1, lty = 2)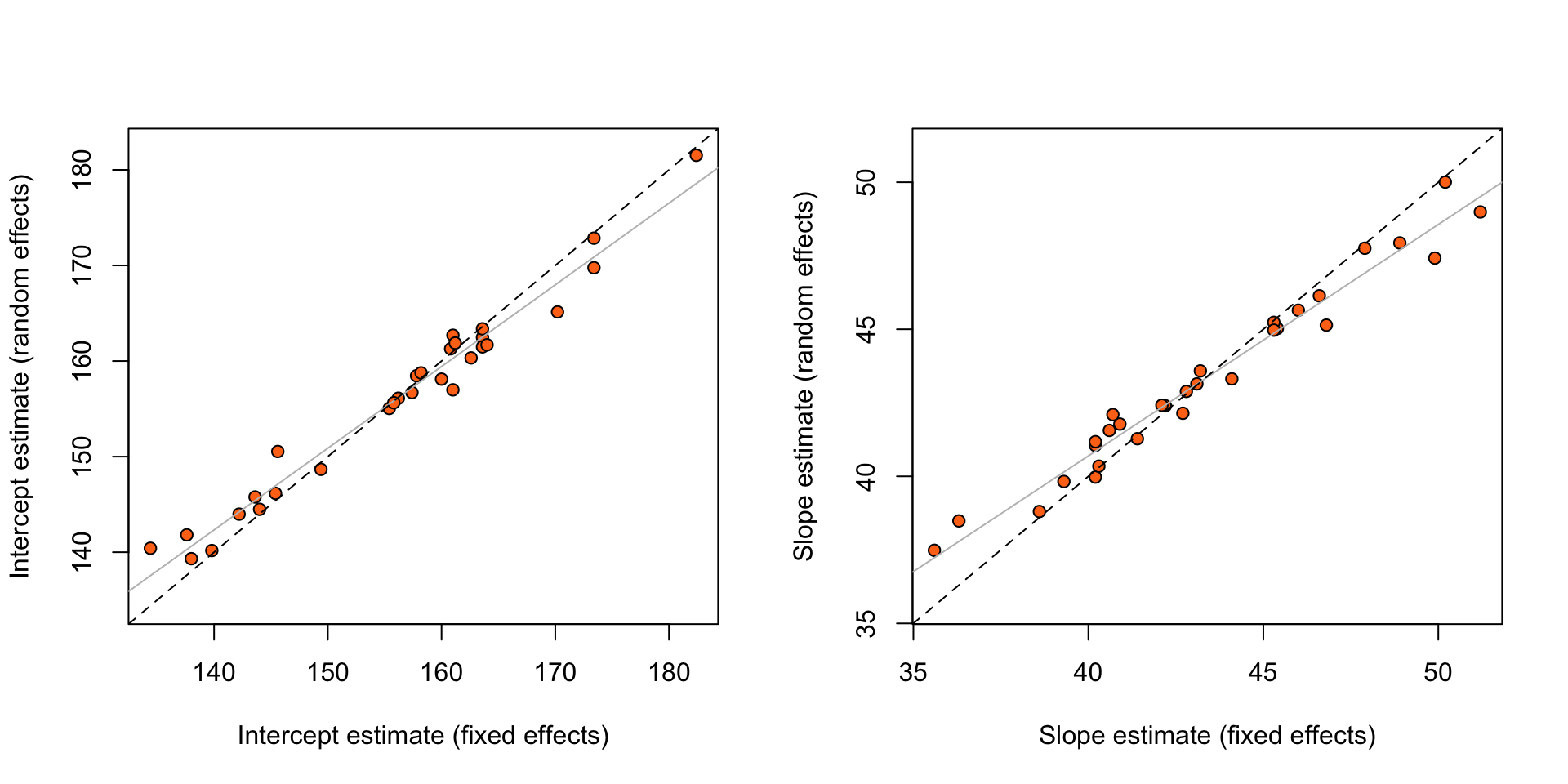
Figure 5: Estimates of random and fixed effects for the rat growth data. The dashed line is a line with intercept zero and slope 1, and the solid line is from the least squares fit of the observed points.
Bayesian inference
Bayesian inference for LMMs relies on Markov Chain Monte Carlo (MCMC) methods, such as the Gibbs sampler, that have proved very effective in practice.
MCMC computation provides posterior summaries, by generating a dependent sample from the posterior distribution of interest. Then, any posterior expectation can be estimated by the corresponding Monte Carlo sample mean, densities can be estimated from samples, etc.
Gibbs sampler
To generate from \(f(y_1, \ldots , y_n)\), where the components \(y_i\) are allowed to be multivariate, the Gibbs sampler
starts at an arbitrary value of \(y\)
updates components by generating from the conditional distributions \(f(y_i \mid y_{\setminus i})\) where \(y_{\setminus i}\) are all the variables other than \(y_i\), set at their currently generated values.
We require conditional distributions which are available for sampling.
Gibbs sampling for LLMs
For the linear mixed model \[ Y \mid X, Z, \beta, \Sigma, b \sim {\mathop{\mathrm{N}}}(\mu, \Sigma),\quad \mu=X\beta+Zb,\quad b \mid \Sigma_b \sim \mathop{\mathrm{N}}(0,\Sigma_b) \] and prior densities \(\pi(\beta)\), \(\pi(\Sigma)\), \(\pi(\Sigma_b)\), we obtain the conditional posterior distributions \[ \begin{aligned} f(\beta\mid y,\text{rest}) & \propto \phi(y-Zb \,;\,X\beta,V)\pi(\beta) \\ f(b\mid y,\text{rest})&\propto \phi(y-X\beta \,;\,Zb,V)\phi(b \,;\,0,\Sigma_b) \\ f(\Sigma\mid y,\text{rest})&\propto \phi(y-X\beta-Zb \,;\,0,V)\pi(\Sigma) \\ f(\Sigma_b\mid y,\text{rest})&\propto \phi(b \,;\,0,\Sigma_b)\pi(\Sigma_b) \end{aligned} \] where \(\phi(y\,;\,\mu,\Sigma)\) is the density of a \({\mathop{\mathrm{N}}}(\mu,\Sigma)\) random variable evaluated at \(y\).
Notes
Exploiting conditional conjugacy in the choices of \(\pi(\beta)\), \(\pi(\Sigma)\), \(\pi(\Sigma_b)\) making the conditionals above of known form and, hence, straightforward to sample from.
Exploiting the conditional independence \((\beta, \Sigma) \perp\!\!\!\perp \Sigma_b \mid b\) is also helpful.
Generalized linear mixed models
Generalized linear mixed models
A generalized linear mixed model (GLMM) has \[ Y_i \mid x_i, z_i, b \stackrel{\text{ind}}{\sim}\mathop{\mathrm{EF}}(\mu_i,\sigma^2)\, , \quad \begin{bmatrix} g(\mu_1)\\\vdots \\ g(\mu_n) \end{bmatrix} = X \beta + Zb \, , \quad b \sim {\mathop{\mathrm{N}}}(0,\Sigma_b) \tag{6}\] where \(\mathop{\mathrm{EF}}(\mu_i,\sigma^2)\) is an exponential family with mean \(\mu_i\) and variance \(\sigma^2 V(\mu_i) / m_i\).
Notes
For many well-used distributions, like binomial and Poisson, \(\sigma^2 = 1\), and we shall assume that from now on, for simplicity of presentation.
The normality of \(b\) can be relaxed in many ways, and to other distributions, but normal random effects usually provide adequate fits.
Example: Random-intercept logistic regression with one covariate
\[ Y_{ij} \mid x_{ij}, b_{i} \stackrel{\text{ind}}{\sim}\mathop{\mathrm{Bernoulli}}(\mu_{ij})\,, \quad \log\frac{\mu_{ij}}{1-\mu_{ij}} = \beta_1 + b_{i} + \beta_2 x_{ij}\,, \quad b_{i} \stackrel{\text{ind}}{\sim}{\mathop{\mathrm{N}}}(0,\sigma^2_b) \]
Suitable, for example, for the toxoplasmosis data.
Fits the general GLMM definition in (6) with \(g(\mu) = \log \{ \mu / (1 - \mu) \}\), and \[ Y = \begin{bmatrix} Y_1 \\ \vdots \\ Y_k \end{bmatrix}\,, \quad Y_i = \begin{bmatrix} Y_{i1} \\ \vdots \\ Y_{in_i} \end{bmatrix} \, , \quad X = \begin{bmatrix} X_1 \\ \cdots \\ X_k \end{bmatrix}\,, \quad X_i = \begin{bmatrix} 1 & x_{i1} \\ \vdots & \vdots \\ 1 & x_{in_i} \end{bmatrix}\,, \] \[ Z = \begin{bmatrix} Z_1 & 0 & \cdots & 0 \\ 0 & Z_2 & \cdots & 0 \\ \vdots & \vdots & \ddots & \vdots \\ 0 & 0 & \cdots & Z_k \end{bmatrix}\,, \quad Z_i = \begin{bmatrix} 1 \\ \vdots \\ 1 \end{bmatrix}\,, \quad \beta = \begin{bmatrix} \beta_1 \\ \beta_2 \end{bmatrix}\,, \quad b = \begin{bmatrix} b_1 \\ \vdots \\ b_k \end{bmatrix}\,, \quad \Sigma_b = \sigma^2_b I_k \]
Likelihood
The marginal distribution of \(Y\) in a GLMM does not usually have a closed-form. \[ \begin{aligned} f(y \mid X, Z \,;\,\beta,\Sigma_b) & = \int f(y \mid X, Z, b \,;\,\beta, \Sigma_b)f(b \,;\,\beta, \Sigma_b)db \\ &= \int f(y \mid X, Z, b \,;\,\beta)f(b \,;\,\Sigma_b)db\\ &= \int \prod_{i=1}^n f\left(y_i \mid X, Z, b \,;\,\beta)\right) f(b \,;\,\Sigma_b)db \end{aligned} \]
For nested random effects structures, some simplification is possible. For example, for the random-intercept logistic regression model in the previous slide \[ f(y \mid X, Z \,;\,\beta,\sigma^2_b) = \prod_{i=1}^k \int \prod_j f \left(y_{ij} \mid x_i, b_i \,;\,\beta)\right) \phi(b_i \,;\,0, \sigma^2_b)db_i \]
\(\Large\Rightarrow\) We need integral approximation methods.
Quadrature
For low-dimensional integrals, a reliable method is to use Gaussian quadrature.
For example, for an one-dimensional cluster-level random effect \(b_i\) \[ \int \prod_j f \left(y_{ij} \mid x_i, b_i \,;\,\beta)\right) \phi(b_i \,;\,0, \sigma^2_b)db_i \approx \sum_{q = 1}^Q W_q \prod_j f \left(y_{ij} \mid x_i, \sqrt{2} \sigma_b B_q \,;\,\beta)\right) / \sqrt{\pi} \] for weights \(W_q\) and quadrature points \(B_q\) \((q = 1,\ldots ,Q)\) chosen according to the Gauss-Hermite quadrature rule.
Notes
Effective quadrature approaches use information about the mode and dispersion of the integrand, which can be done adaptively.
For multi-dimensional \(b_i\), quadrature rules can be applied recursively, but performance in fixed-time diminishes rapidly with dimension.
Laplace approximation
An alternative approach is to use a Laplace approximation to the likelihood.
Writing, \[ h(b) = \prod_{i=1}^n f\left(y_i \mid X, Z, b \,;\,\beta\right) f(b \,;\,\Sigma_b) \] for the integrand of the likelihood, a first-order Laplace approximation approximates \(h(.)\) as an unnormalized multivariate normal density function \[ \tilde h(b) = c \, \phi_k(b \,;\,\hat b, Q) \]
\(\hat b\) is found by maximizing \(\log h(.)\) over \(b\)
the variance matrix \(Q\) is chosen so that the curvature of \(\log h(.)\) and \(\log \tilde h(.)\) agree at \(\hat b\)
\(c\) is chosen so that \(\tilde h(\hat b) = h(\hat b)\)
Laplace approximation
Notes
The first-order Laplace approximation is equivalent to adaptive Gaussian quadrature with a single quadrature point.
The accuracy of Laplace approximation is not guaranteed for every model.
Estimation of GLMMs
Estimation and inference from GLMMs remains an area of active research and vigorous debate.
Other approaches include:
Penalized quasi-likelihood (PQL), which is based on a normal approximation of the distribution of the adjusted dependent variables when computing maximum likelihood estimators for GLMs using Fisher scoring.
Hierarchical GLMs (HGLMs) where inference is based on the h-likelihood \(f(y \mid X, Z, b \,;\,\beta, \Sigma_b)f(b \,;\,\beta, \Sigma_b)\).
There are cases where both these approaches work well, but also cases where they can fail badly, so they should be used with caution.
Model comparison should not be performed with the PQL and HGLM objective functions.
Toxoplasmosis
Fitting a logistic regression with city-specific random intercepts and a linear relationship with rainfall (divided by \(10^5\)) gives
library("MASS")
library("lme4")
library("modelsummary")
toxo$city <- 1:nrow(toxo)
toxo$rain_s <- toxo$rain / 100000
mod_lmm_quad <- glmer(r/m ~ rain_s + (1 | city), weights = m,
data = toxo, family = binomial, nAGQ = 25)
mod_lmm_LA <- glmer(r/m ~ rain_s + (1 | city), weights = m,
data = toxo, family = binomial)
mod_lmm_PQL <- glmmPQL(fixed = r/m ~ rain_s, random = ~ 1 | city, weights = m,
data = toxo, family = binomial)
modelsummary(
list("GH (25)" = mod_lmm_quad,
"Laplace" = mod_lmm_LA,
"PQL" = mod_lmm_PQL),
output = "markdown",
escape = FALSE,
gof_map = "none",
fmt = fmt_decimal(digits = 3),
coef_omit = 4,
coef_rename = c("$\\beta_1$", "$\\beta_2$", "$\\sigma_b$"))| GH (25) | Laplace | PQL | |
|---|---|---|---|
| \(\beta_1\) | -0.138 | -0.134 | -0.115 |
| (1.450) | (1.441) | (1.445) | |
| \(\beta_2\) | 0.722 | 0.592 | 0.057 |
| (75.060) | (74.620) | (74.922) | |
| \(\sigma_b\) | 0.521 | 0.513 | 0.495 |
Toxoplasmosis
Fitting a logistic regression with city-specific random intercepts and a cubic relationship with rainfall (divided by \(10^5\)) gives
mod_lmm_cubic_quad <- glmer(r/m ~ poly(rain_s, 3) + (1 | city), weights = m,
data = toxo, family = binomial, nAGQ = 25)
mod_lmm_cubic_LA <- glmer(r/m ~ poly(rain_s, 3) + (1 | city), weights = m,
data = toxo, family = binomial)
mod_lmm_cubic_PQL <- glmmPQL(fixed = r/m ~ poly(rain_s, 3), random = ~ 1 | city, weights = m,
data = toxo, family = binomial)
modelsummary(
list("GH (25)" = mod_lmm_cubic_quad,
"Laplace" = mod_lmm_cubic_LA,
"PQL" = mod_lmm_cubic_PQL),
output = "markdown",
escape = FALSE,
gof_map = "none",
fmt = fmt_decimal(digits = 3),
coef_omit = 6,
coef_rename = c("$\\beta_1$", "$\\beta_2$", "$\\beta_3$", "$\\beta_4$", "$\\sigma_b$"))| GH (25) | Laplace | PQL | |
|---|---|---|---|
| \(\beta_1\) | -0.106 | -0.104 | -0.110 |
| (0.127) | (0.126) | (0.127) | |
| \(\beta_2\) | -0.106 | -0.107 | -0.098 |
| (0.687) | (0.682) | (0.718) | |
| \(\beta_3\) | 0.154 | 0.149 | 0.173 |
| (0.700) | (0.695) | (0.724) | |
| \(\beta_4\) | 1.628 | 1.626 | 1.607 |
| (0.654) | (0.649) | (0.682) | |
| \(\sigma_b\) | 0.423 | 0.417 | 0.431 |
Toxoplasmosis
The AIC and BIC (using \(25\) Gauss-Hermite quadrature points for the likelihood approximation) for the linear and the cubic individual-level models are
Data: toxo
Models:
mod_lmm_quad: r/m ~ rain_s + (1 | city)
mod_lmm_cubic_quad: r/m ~ poly(rain_s, 3) + (1 | city)
npar AIC BIC logLik deviance Chisq Df Pr(>Chisq)
mod_lmm_quad 3 65.754 70.333 -29.877 59.754
mod_lmm_cubic_quad 5 63.840 71.472 -26.920 53.840 5.9139 2 0.05198 .
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Bayesian inference
As for LMMs, Bayesian inference in GLMMs is typically based on the Gibbs sampler.
For prior densities \(\pi(\beta)\) and \(\pi(\Sigma_b)\) and known \(\sigma^2\), we obtain the conditional posterior distributions \[ \begin{aligned} f(\beta\mid y,\text{rest}) & \propto \pi(\beta) \prod_i f(y_i \mid X, Z, \beta, b)\\ f(b\mid y,\text{rest}) & \propto \phi(b \,;\,0, \Sigma_b)\prod_i f(y_i \mid X, Z, \beta, b)\\ f(\Sigma_b \mid y, \text{rest}) & \propto \phi(b \,;\,0, \Sigma_b) \pi(\Sigma_b) \end{aligned} \]
For a conditionally conjugate choice of \(\pi(\Sigma_b)\), \(f(\Sigma_b \mid y, \text{rest})\) is easy to sample from.
The conditionals for \(\beta\) and \(b\) are not generally available for direct sampling. However, there are a number of ways of modifying the basic Gibbs sampling to go around this.
| Materials | Link |
|---|---|
| Preliminary material | ikosmidis.com/files/APTS-SM-Preliminary |
| Notes | ikosmidis.com/files/APTS-SM-Notes |
| Slides: Introduction | ikosmidis.com/files/APTS-SM-Slides-intro |
| Slides: Model selection | ikosmidis.com/files/APTS-SM-Slides-model-selection |
| Slides: Beyond GLMs | ikosmidis.com/files/APTS-SM-Slides-beyond-glms |
| Slides: Nonlinear models | ikosmidis.com/files/APTS-SM-Slides-nonlinear-models |
| Slides: Latent variables | ikosmidis.com/files/APTS-SM-Slides-latent |
| Lab 1 | ikosmidis.com/files/APTS-SM-Notes/lab1.html |
| Lab 2 | ikosmidis.com/files/APTS-SM-Notes/lab2.html |

Ioannis Kosmidis - Statistical Modelling: Beyond GLMs

