'data.frame': 53 obs. of 7 variables:
$ m : Named num 1 1 1 1 1 1 1 1 1 1 ...
..- attr(*, "names")= chr [1:53] "1" "2" "3" "4" ...
$ r : Named num 1 1 1 1 1 0 1 0 0 0 ...
..- attr(*, "names")= chr [1:53] "1" "2" "3" "4" ...
$ aged : Factor w/ 2 levels "0","1": 1 1 1 1 1 1 1 1 1 1 ...
..- attr(*, "names")= chr [1:53] "1" "2" "3" "4" ...
$ stage: Factor w/ 2 levels "0","1": 2 2 2 2 2 2 1 1 1 1 ...
..- attr(*, "names")= chr [1:53] "1" "2" "3" "4" ...
$ grade: Factor w/ 2 levels "0","1": 2 2 2 2 2 2 1 1 1 1 ...
..- attr(*, "names")= chr [1:53] "1" "2" "3" "4" ...
$ xray : Factor w/ 2 levels "0","1": 2 2 2 2 2 2 1 1 1 1 ...
..- attr(*, "names")= chr [1:53] "1" "2" "3" "4" ...
$ acid : Factor w/ 2 levels "0","1": 2 2 2 2 2 2 2 2 2 2 ...
..- attr(*, "names")= chr [1:53] "1" "2" "3" "4" ...APTS Statistical Modelling
Model Selection
Professor of Statistics
Department of Statistics, University of Warwick
ioannis.kosmidis@warwick.ac.uk
ikosmidis.com ikosmidis ikosmidis_
19 April 2024
Model selection
Task
Select a statistical model from a set of candidate models on the basis of data.
Principles for selection
Background knowledge
Choose models based on substantive knowledge (e.g. from previous studies), theoretical arguments, dimensional or other general considerations.
Robustness to departures from assumptions
Prefer models that provide valid inference even if some of their assumptions are invalid.
Quality of fit
Prefer models that perform well in terms of informal devices such as residuals and graphical assessments, or more formal or goodness-of-fit tests.
Parsimony
Look for the simplest models that are adequate descriptions of the data.
Set of candidate models
Defined based on background knowledge, mathematical or computational convenience.
Defined accounting for the trade-off between allowed flexibility and cardinality of the set of candidate models.
Linear regression
Data
Response values \(y_1, \ldots, y_n\), with \(y_i \in \Re\)
Covariate vectors \(x_1, \ldots, x_n\), with \(x_i = (x_{i1}, \ldots, x_{ip})^\top \in \Re^p\)
Model
\[ Y_i = \sum_{t = 1}^p \beta_t x_{it} + \epsilon_i \] where \(\mathop{\mathrm{E}}(\epsilon_i) = 0\) and \(\mathop{\mathrm{var}}(\epsilon_i) = \sigma^2 < \infty\)
Set of candidate models
For \(p = 30\), assuming that every model has an intercept parameter (\(x_{i1} = 1\)), the set of candidate models has about 537 million possible models, without even accounting for covariate transformations or interactions!
Logistic regression
Data
Response values \(y_1, \ldots, y_n\), with \(y_i \in \{0, 1\}\)
Covariate vectors \(x_1, \ldots, x_n\), with \(x_i = (x_{i1}, \ldots, x_{ip})^\top \in \Re^p\)
Model
\(Y_{1}, \ldots, Y_{n}\) conditionally independent with
\[ Y_i \mid x_i \sim \mathop{\mathrm{Bernoulli}}(\mu_i)\,, \quad \log \frac{\mu_i}{1 - \mu_i} = \eta_i = \sum_{t = 1}^p \beta_t x_{it} \]
Widely used for inference about covariate effects on probabilities, probability calibration, and prediction.
Set of candidate models
Same considerations as for linear regression.
Logistic regression likelihood
\[ \ell(\beta) = \sum_{i=1}^n y_i x_i^\top \beta - \sum_{i=1}^n \log\left\{1 + \exp( x_i^\top \beta)\right\} \]
We expect a good fit to be one that gives large maximized log-likelihood \(\hat \ell = \ell(\hat\beta)\) where \(\hat\beta\) is the maximum likelihood estimator
Nodal involvement: ?SMPracticals::nodal
The treatment strategy for a patient diagnosed with cancer of the prostate depend highly on whether the cancer has spread to the surrounding lymph nodes.
It is common to operate on the patient to get samples from the nodes which are then analysed under a microscope.
For a sample of 53 prostate cancer patients, a number of possible predictor variables were measured before surgery. The patients then had surgery to determine nodal involvement.
Nodal involvement
Considering only the models with an intercept and without any interaction between the 5 binary covariates, results in \(2^5 = 32\) possible logistic regression models for this data.
library("SMPracticals")
library("MuMIn")
nodal_full <- glm(r ~ aged + stage + grade + xray + acid,
data = nodal, family = "binomial", na.action = "na.fail")
nodal_table <- dredge(nodal_full, rank = logLik)
plot(logLik ~ df, data = nodal_table,
xlab = "Number of parameters",
ylab = "Maximized log-likelihood",
bg = "#ff7518", pch = 21)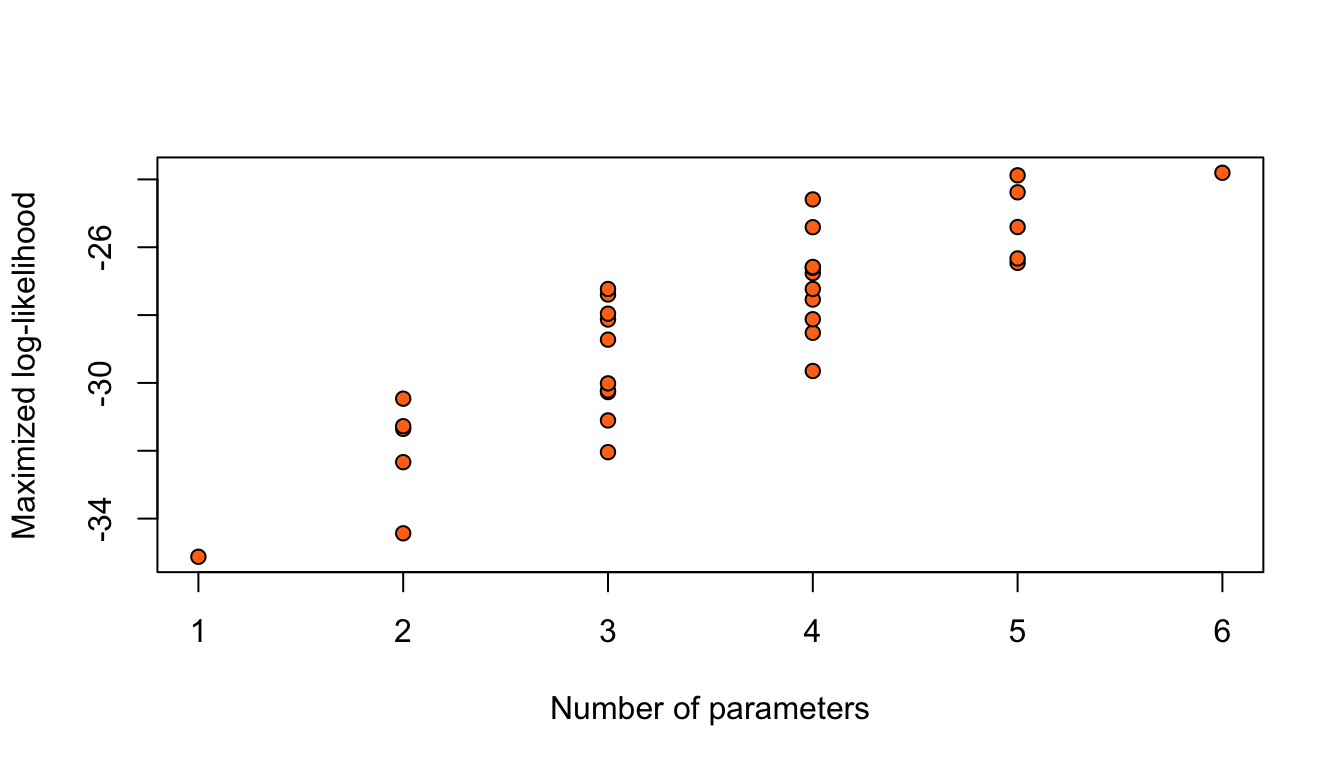
Figure 1: Maximized log-likelihoods for 32 possible logistic regression models for the nodal data.
Adding terms always increases the maximized log-likelihood.
Taking the model with highest \(\hat\ell\) would give the full model.
Linear regression
Let \(Y_i = \eta_i + \epsilon_i\). Removing terms always increases the residual sums of squares \(\|y - \hat{\eta} \|_2^2\).
Criteria
Setting
True model
Independent \(Y_1, \ldots, Y_n\), where \(Y_i\) has a density or probability mass function \(g(y)\).
Candidate model
Independent \(Y_1, \ldots, Y_n\), where \(Y_i\) has a density or probability mass function \(f(y \,;\,\theta)\).
There may be no value of \(\theta\) such that \(f(\cdot \,;\,\theta) = g(\cdot)\).
Log-likelihood
Jensen’s inequality implies \[ \int \log g(y) g(y) dy \ge \int \log f(y \,;\,\theta) g(y) dy \tag{1}\] with equality if and only if \(f(y \,;\,\theta) = g(y)\).
If \(\theta_g\) is the value that maximizes the expectation in the right hand side of (1), then it is natural to choose the candidate model that maximizes the estimate of that expectation, i.e. \[ \bar\ell(\hat\theta) = n^{-1} \sum_{i = 1}^n \log f(y_i \,;\,\hat\theta) \]
However, by definition of \(\hat\theta\), \(\bar\ell(\hat\theta) \ge \bar\ell(\theta_g)\). So, \(\bar\ell(\hat\theta)\) is biased upwards.
Kullback-Leibler divergence
The log-likelihood \(\ell(\theta)\) of the candidate model is maximized at \(\hat\theta\), and \[ \bar \ell(\hat\theta) = n^{-1}\ell(\hat\theta) \to \int \log f(y \,;\,\theta_g) g(y)\, dy, \quad \text{as } n\to\infty \] where \(\theta_g\) minimizes the Kullback-Leibler divergence \[ KL(f_\theta,g) = \int \log\left\{ \frac{g(y)}{f(y \,;\,\theta)}\right\} g(y) \, dy \]
\(\theta_g\) gives the density \(f(y \,;\,\theta_g)\) closest to \(g(y)\), in this sense.
Inference under model misspecification
Asymptotic distribution of \(\hat\theta\)
Under mild regularity conditions, the maximum likelihood estimator \(\hat\theta\) satisfies \[ \sqrt{n} (\hat\theta - \theta_g) \stackrel{d}{\rightarrow} {\mathop{\mathrm{N}}}\left( 0, n I(\theta_g)^{-1}K(\theta_g)I(\theta_g)^{-1}\right) \tag{2}\] where \[ \begin{aligned} K(\theta) &= n \int \frac{\partial \log f(y \,;\,\theta)}{\partial\theta} \frac{\partial \log f(y \,;\,\theta)}{\partial\theta^\top }g(y)\, dy,\\ I(\theta) &= -n \int \frac{\partial^2 \log f(y \,;\,\theta)}{\partial\theta\partial\theta^\top } g(y)\, dy \end{aligned} \]
Inference under model misspecification
Likelihood ratio statistic
The likelihood ratio statistic converges in distribution as \[ W(\theta_g)=2\left\{\ell(\hat\theta)-\ell(\theta_g)\right\} \stackrel{d}{\rightarrow} \sum_{r=1}^p \lambda_r V_r \] where \(V_1, \ldots, V_p\) are independent with \(V_i \sim \chi^2_1\), and \(\lambda_r\) are eigenvalues of \(K(\theta_g)^{1/2}I(\theta_g)^{-1} K(\theta_g)^{1/2}\).
Thus, \(E\{W(\theta_g)\} \rightarrow \mathop{\mathrm{tr}}\{I(\theta_g)^{-1}K(\theta_g)\}\).
Inference under model misspecification
Estimating \(K(\theta_g)\) and \(I(\theta_g)\)
In practice \(g(y)\) is, of course, unknown, and \(K(\theta_g)\) and \(I(\theta_g)\) are estimated by \[ \hat K = \sum_{i=1}^n \frac{\partial \log f(y_i \,;\,\hat\theta)}{\partial\theta} \frac{\partial \log f(y_i \,;\,\hat\theta)}{\partial\theta^\top }, \quad \underbrace{\hat J = - \sum_{i=1}^n \frac{\partial^2 \log f(y_i \,;\,\hat\theta)} {\partial\theta\partial\theta^\top }}_{\text{observed information}} \]
Asymptotically valid inference
We can construct inferences about \(\theta_g\), using the fact that the approximate distribution of \(\hat\theta\) is \({\mathop{\mathrm{N}}}\left( \theta_g, I(\theta_g)^{-1}K(\theta_g)I(\theta_g)^{-1}\right)\), replacing the variance covariance matrix with \(\hat J^{-1}\hat K\hat J^{-1}\).
Maximum likelihood asymptotics under the true model
If \(g(\theta) = f(y \,;\,\theta_g)\), \(K(\theta) = I(\theta)\) and \(\lambda_1 = \ldots = \lambda_p = 1\), and we recover the usual results.
Out-of-sample prediction
Problems with using \(\bar\ell(\hat\theta)\) for model selection:
- Upward bias, as \(\bar\ell(\hat\theta) \ge \bar\ell(\theta_g)\) because \(\hat\theta\) is based on \(Y_1, \ldots, Y_n\)
- Improves quality of fit with no control over model complexity
If we had another independent sample \(Y_1^+,\ldots,Y_n^+\sim g\) and computed \[ \bar\ell^+(\hat\theta) = n^{-1}\sum_{i=1}^n \log f(Y^+_i \,;\,\hat\theta) \] then both problems disappear. In the limit, we choose the candidate model that maximizes \[ \Delta = {\mathop{\mathrm{E}}}_g\left[ {\mathop{\mathrm{E}}}^+_g\left\{ \bar\ell^+(\hat\theta) \right\} \right] \] where the inner expectation is over the distribution of \(Y_i^+\), and the outer expectation is over the distribution of \(\hat\theta\).
Bias of \(\bar \ell(\hat \theta)\) as an estimator of \(\Delta\)
\[ {\mathop{\mathrm{E}}}_g\{\bar \ell(\hat \theta)\} - \Delta = \underbrace{{\mathop{\mathrm{E}}}_g\{\bar \ell(\hat \theta) - \bar \ell(\theta_g)\}}_{a} + \underbrace{{\mathop{\mathrm{E}}}_g\{\bar \ell(\theta_g)\} - \Delta}_{b} \]
From the asymptotic distribution of \(W(\theta_g)\) \[ \begin{aligned} a & = {\mathop{\mathrm{E}}}_g\{\bar\ell(\hat\theta) - \bar\ell(\theta_g)\} \\ & = \frac{1}{2n}E_g\{W(\theta_g)\} \\ & \approx \frac{1}{2n}\mathop{\mathrm{tr}}\{I(\theta_g)^{-1} K(\theta_g)\} \end{aligned} \]
A Taylor expansion of \(\bar\ell^+(\hat\theta)\) around \(\theta_g\) and results on inference under the wrong model can be used to show that \[ \begin{aligned} b & = {\mathop{\mathrm{E}}}_g\{\bar \ell(\theta_g)\} - \Delta \\ & \approx \frac{1}{2n} \mathop{\mathrm{tr}}\{I(\theta_g)^{-1} K(\theta_g)\} \end{aligned} \]
\[\LARGE\Downarrow\]
\[ {\mathop{\mathrm{E}}}_g\{\bar \ell(\hat \theta)\} - \Delta \approx \frac{1}{n} \mathop{\mathrm{tr}}\{I(\theta_g)^{-1} K(\theta_g)\} \]
Takeuchi information criterion
We can correct the bias of \(\bar\ell(\hat\theta)\) as an estimator \(\Delta\), by maximizing \[ \bar\ell(\hat\theta) - \frac{1}{n}\mathop{\mathrm{tr}}(\hat J^{-1} \hat K) \] over the candidate models.
Equivalent formulations
Maximize \[ \hat \ell - \mathop{\mathrm{tr}}(\hat J^{-1} \hat K) \]
Minimize \[ 2\{\mathop{\mathrm{tr}}(\hat J^{-1}\hat K)-\hat\ell\} \tag{3}\]
(3) is called the Takeuchi Information Criterion and has also been refereed to as the Network Information Criterion.
Information criteria
| Information criterion | Acronym | Definition |
|---|---|---|
| Takeuchi Information Criterion | TIC | \(2\{\mathop{\mathrm{tr}}(\hat J^{-1}\hat K) - \hat\ell\}\) |
| Akaike Information Criterion | AIC | \(2(p - \hat\ell)\) |
| Corrected AIC | AIC\(_\text{c}\) | \(2(p + (p^2 + p) / (n - p - 1) - \hat\ell)\) |
| Bayesian Information Criterion | BIC | \(2(p \log n / 2 - \hat\ell)\) |
| Deviance Information Criterion | DIC | |
| Extended Information Criterion | EIC | |
| Generalized Information Criterion | GIC | |
| \(\vdots\) |
A popular model selection criterion in regression problems is Mallows’ \(C_p = \text{RSS}/s^2 + 2p-n\), where \(s^2\) is an estimate of the error variance \(\sigma^2\).
Nodal involvement: AIC
Global model call: glm(formula = r ~ aged + stage + grade + xray + acid, family = "binomial",
data = nodal, na.action = "na.fail")
---
Model selection table
(Intrc) acid aged grade stage xray df logLik AIC delta weight
26 -3.052 + + + 4 -24.590 57.2 0.00 0.268
30 -3.262 + + + + 5 -23.880 57.8 0.58 0.201
28 -2.778 + + + + 5 -24.380 58.8 1.58 0.122
22 -2.734 + + + 4 -25.409 58.8 1.64 0.118
32 -3.079 + + + + + 6 -23.805 59.6 2.43 0.080
25 -2.082 + + 3 -27.231 60.5 3.28 0.052
18 -2.176 + + 3 -27.394 60.8 3.61 0.044
24 -2.687 + + + + 5 -25.404 60.8 3.63 0.044
14 -2.894 + + + 4 -26.581 61.2 3.98 0.037
27 -1.735 + + + 4 -26.606 61.2 4.03 0.036
Models ranked by AIC(x) Nodal involvement: BIC
Global model call: glm(formula = r ~ aged + stage + grade + xray + acid, family = "binomial",
data = nodal, na.action = "na.fail")
---
Model selection table
(Intrc) acid aged grade stage xray df logLik BIC delta weight
26 -3.052 + + + 4 -24.590 65.1 0.00 0.278
25 -2.082 + + 3 -27.231 66.4 1.31 0.144
22 -2.734 + + + 4 -25.409 66.7 1.64 0.123
18 -2.176 + + 3 -27.394 66.7 1.64 0.123
30 -3.262 + + + + 5 -23.880 67.6 2.55 0.078
10 -2.509 + + 3 -27.957 67.8 2.76 0.070
6 -2.418 + + 3 -28.131 68.2 3.11 0.059
28 -2.778 + + + + 5 -24.380 68.6 3.55 0.047
17 -1.135 + 2 -30.465 68.9 3.81 0.041
14 -2.894 + + + 4 -26.581 69.0 3.98 0.038
Models ranked by BIC(x) Nodal involvement: AIC, BIC
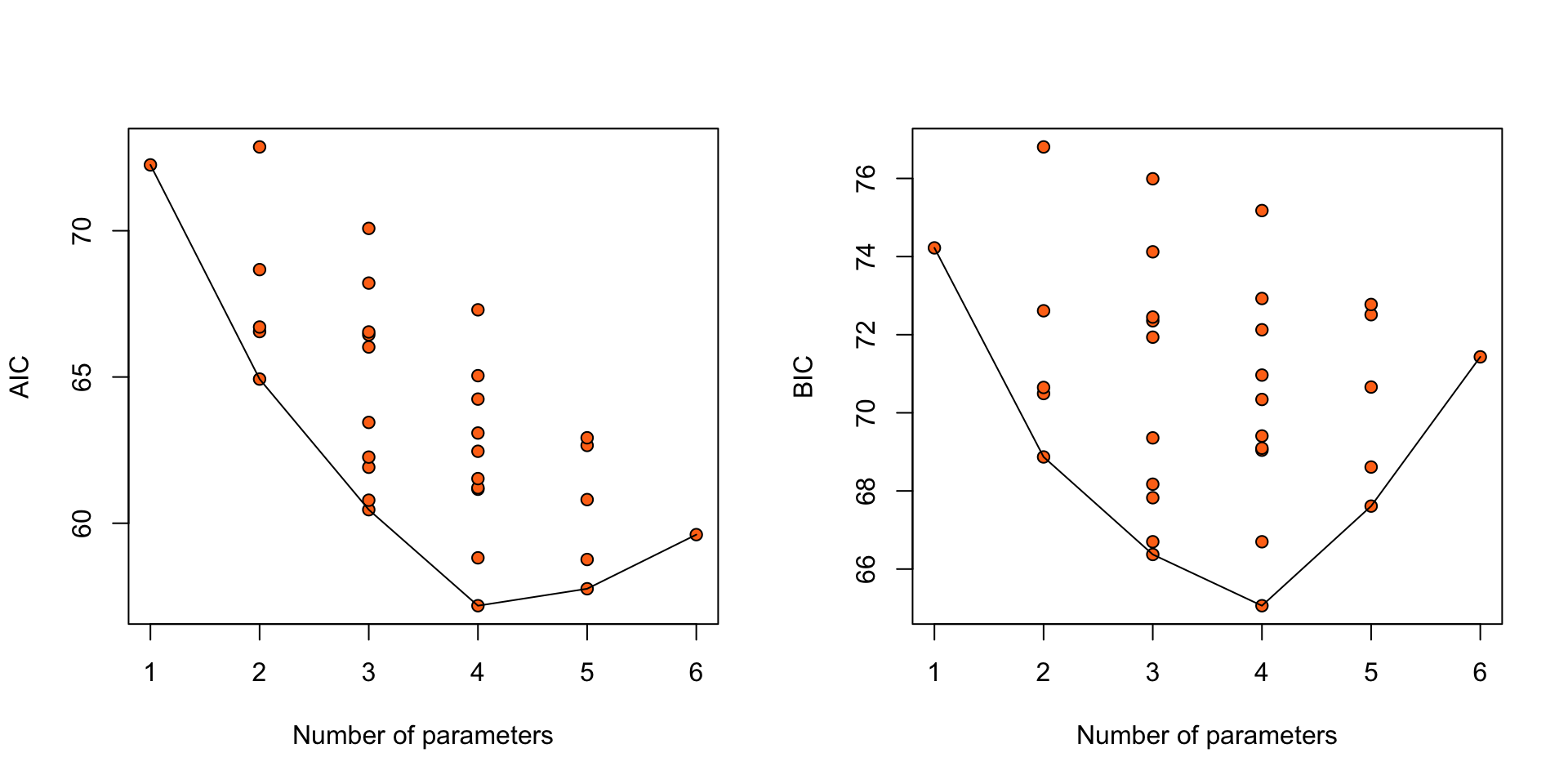
Figure 2: AIC and BIC for 32 logistic regression models for the nodal data.
Properties of model selection procedures
Asymptotic efficiency
Suppose the true model is of infinite dimensions. A model selection procedure that selects the candidate “closest” to the true model for large \(n\) is called asymptotically efficient.
Consistency
Suppose that the true model is among the candidate models. A model selection procedure that selects the true model with probability tending to one as \(n\to\infty\) is called consistent.
Asymptotic analysis of AIC, BIC, TIC
Let \(\text{IC} = c(n, p) - 2\hat\ell\) be an information criterion for a candidate model, and IC\(_+\) be the value of the information criterion for a model with one extra parameter.
\[ \begin{aligned} P(\text{IC}_+ < \text{IC}) &= P\left\{ c(n,p+1)-2\hat\ell_+< c(n,p)-2\hat\ell\right\} \\ & = P\left\{2(\hat\ell_+ - \hat\ell) > d(n, p) \right\} \end{aligned} \]
| Criterion | \(d(n, p)\) |
|---|---|
| AIC | \(= 2\) |
| TIC | \(\approx 2\) for large \(n\) |
| BIC | \(= \log n\) |
\(2(\hat\ell_+-\hat\ell)\) converges in distribution to a \(\chi^2_1\) random variable. So, as \(n\to\infty\), \[ P(\text{IC}_+<\text{IC}) \to \begin{cases} 0.157, & \text{if AIC or TIC is used} \\ 0, & \text{if BIC is used} \end{cases} \]
Linear models
Variable selection for linear models
Model
\(Y = X^*\beta + \epsilon\)
\(\mathop{\mathrm{E}}(\epsilon) = 0\), \(\mathop{\mathrm{cov}}(\epsilon) = \sigma^2 I_n\)
\(X^*\) is an \(n \times p^*\) matrix with rows \(x_1^*, \ldots, x_n^*\) and \(j\)th column \(v_j\), \(j \in \mathcal{I}= \{1, \ldots, p^*\}\)
Data generating process
\(Y = X^\dagger \beta^\dagger + \epsilon\)
\(X^\dagger\) is an \(n \times p_0\) matrix with columns \(\{v_j : j \in \mathcal{J}\subseteq \mathcal{I}\}\), \(|\mathcal{J}| = p_0 \le p^*\)
Aim
Estimate \(\mathcal{J}\) based on data
Variable selection for linear models
Column selection is variable selection
Selecting columns of \(X^*\) is equivalent to estimating the sets \[ \mathcal{J}= \{j \in \mathcal{I}: \beta_j \ne 0\} \quad \text{and} \quad \mathcal{K}= \{j \in \mathcal{I}: \beta_j = 0\} \]
Candidate model
\(Y = X\beta + \epsilon\)
\(X\) has the columns of \(X^*\) with indices in \(\mathcal{S}\subseteq \mathcal{I}\), \(|\mathcal{S}| = p \le p^*\)
Types of selected models
| Type | Selected set \(\mathcal{S}\) |
|---|---|
| Correct | \(\mathcal{J}\subseteq \mathcal{S}\subseteq \mathcal{I}\) |
| Wrong | \(\mathcal{S}\not\subset \mathcal{J}\) |
| True | \(\mathcal{J}\) |
Prediction mean squared error
Fitted values from candidate model
\[ X\hat\beta = X \{ (X^\top X)^{-1}X^\top Y\} = H Y = H\mu + H\epsilon \]
\(\mu = \mathop{\mathrm{E}}(Y \mid X^*) = X^* \beta\)
\(H = X (X^\top X)^{-1} X^\top\) is the hat matrix
Prediction mean squared error
\[ \Delta = n^{-1} {\mathop{\mathrm{E}}} \left[{\mathop{\mathrm{E}}}_+\left\{ (Y_+ - X\hat\beta)^\top (Y_+ - X\hat\beta) \right\}\right] \] where expectations are taken over both \(Y\) and \(Y_+\)
\(Y_+ = \mu + \epsilon_+\) with \(\epsilon_+\) is an independent copy of \(\epsilon\)
Bias / Variance trade-off
\(Y_{+} - X\hat\beta = {\color[rgb]{1,1,1}{Y_{+} - H Y = \mu + \epsilon_{+} - H \mu - H \epsilon = (I_n - H) \mu + (\epsilon_{+} - H \epsilon)}}\)
\((Y_{+} - X\hat\beta)^\top (Y_{+} - X\hat\beta) = {\color[rgb]{1,1,1}{\mu (I_n - H) \mu + \epsilon_{+}^\top \epsilon_{+} + \epsilon^\top H \epsilon + Z}}\)
where \(Z\) collects all terms with \({\mathop{\mathrm{E}}}[{\mathop{\mathrm{E}}}_{+}(Z)] = 0\). From the assumptions on the errors, we have \({\mathop{\mathrm{E}}}[{\mathop{\mathrm{E}}}_{+}(\epsilon_{+}^\top \epsilon_{+})] = n \sigma^2\), and \({\mathop{\mathrm{E}}}[{\mathop{\mathrm{E}}}_{+}(\epsilon^\top H \epsilon)] = \mathop{\mathrm{tr}}{H}\sigma^2 = p\sigma^2\). Collecting terms
Prediction mean squared error
\[ \Delta = \underbrace{\mu (I_n - H) \mu / n}_{\text{Bias}} + \overbrace{(1 + p / n) \sigma^2}^{\text{Variance}} \]
Bias / Variance trade-off
| Type | Selected set \(\mathcal{S}\) | \(\Delta\) |
|---|---|---|
| Wrong | \(\mathcal{S}\not\subset \mathcal{J}\) | \(\mu (I_n - H) \mu / n + (1 + p / n) \sigma^2\) |
| Correct | \(\mathcal{J}\subseteq \mathcal{S}\subseteq \mathcal{I}\) | \((1 + p / n) \sigma^2\), \(p \ge p_0\) |
| True | \(\mathcal{J}\) | \((1 + p_0 / n) \sigma^2\) |
Bias
\(n^{-1}\mu^\top (I_n - H) \mu = n^{-1}\|\mu - H\mu\|_2^2 \ge 0\), and zero only if the model is correct (\(H\mu = \mu\))
Bias is reduced the closer \(\mu\) is to the space spanned by the columns of \(X\)
We expect a reduction in bias when useful covariates are added to the model
Variance
\((1 + p/n) \sigma^2\) increases as \(p\) increases, for example whenever useless terms are included
Polynomial regression
True model
\[ Y_i = \sum_{j = 0}^{5} x_i^j + \epsilon_i \] \(\mu_i = \sum_{j = 0}^{5} x_i^j\) is a degree five polynomial
Candidate models
\[ Y_i = \sum_{j = 0}^{p - 1} \beta_j x_i^j + \epsilon_i \quad (i = 1, \ldots, n)\, , \]
\(x_1, \ldots, x_n\) generated from \(n\) independent \(\mathop{\mathrm{N}}(0, 1)\)
Polynomial regression
\(n = 20\) and \(\sigma^2 = 1\)
Delta <- function(p, p0, x, sigma2) {
cols <- 0:(p - 1)
cols0 <- 0:(p0 - 1)
n <- length(x)
X <- matrix(rep(x, p)^rep(cols, each = n), nrow = n)
X0 <- matrix(rep(x, p0)^rep(cols0, each = n), nrow = n)
mu <- rowSums(X0)
H <- tcrossprod(qr.Q(qr(X)))
bias <- sum(((diag(n) - H) %*% mu)^2) / n
variance <- sigma2 * (1 + p / n)
c(p = p, bias = bias, variance = variance)
}
n <- 20
sigma2 <- 1
p_max <- 15
set.seed(1)
x <- rnorm(n)
D <- data.frame(t(sapply(1:p_max, Delta, p0 = 6, x = x, sigma2 = 1)))
par(mfrow = c(1, 3))
plot(sqrt(bias) ~ p, data = D, subset = p > 2,
main = expression(sqrt("Bias")), ylab = "",
type = "b", bg = "#ff7518", pch = 21)
abline(v = 6, lty = 3, col = "grey")
plot(sqrt(variance) ~ p, data = D, subset = p > 2,
main = expression(sqrt("Variance")), ylab = "",
type = "b", bg = "#ff7518", pch = 21)
abline(v = 6, lty = 3, col = "grey")
plot(sqrt(bias + variance) ~ p, data = D, subset = p > 2,
main = expression(sqrt(Delta)), ylab = "",
type = "b", bg = "#ff7518", pch = 21)
abline(v = 6, lty = 3, col = "grey")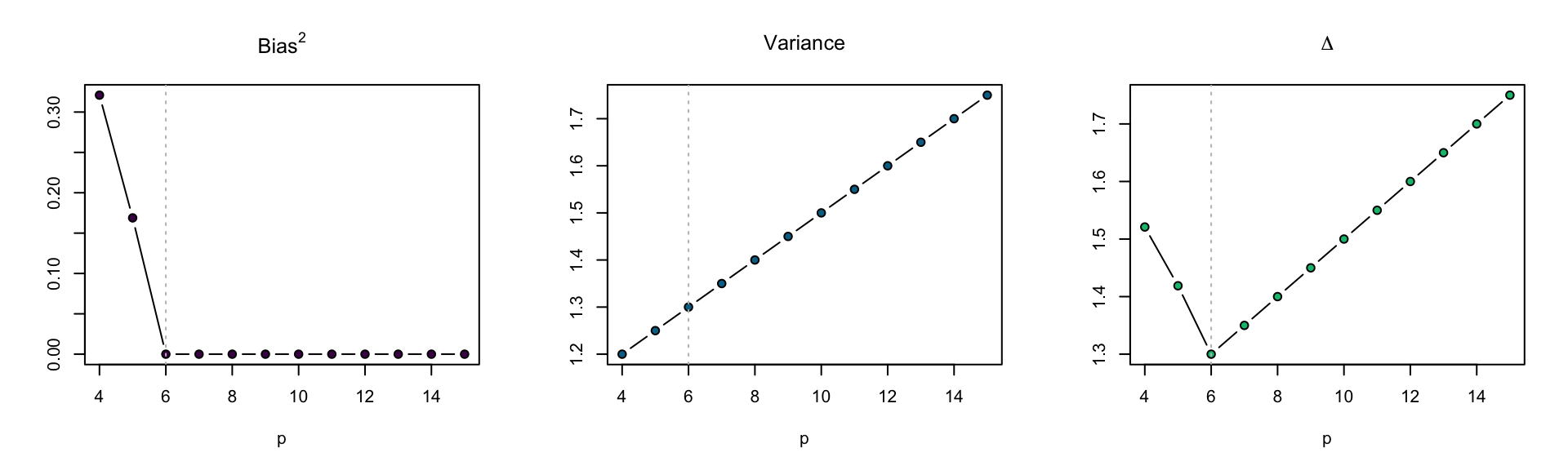
Figure 3: \(\Delta\) for models with varying polynomial degree.
Data splitting
\(\Delta\) depends on \(\mu\) and \(\sigma^2\), which are unknown. But we can use the assumption of exchangeability of \(Y_1, \ldots, Y_n\) given the covariates to estimate it.
For each candidate model:
Split data into \((X, y)\) and \((X_{+}, y_{+})\), where \(X_{+}\) is an \(n_{+} \times p\) matrix, and \(y_{+}\) is the vector of the corresponding \(n_{+}\) response values.
Estimate the model using \((X, y)\).
Compute \[ \hat\Delta = \frac{1}{n_{+}}(y_{+} - X_{+}\hat\beta)^\top (y_{+} - X_{+}\hat\beta) = \frac{1}{n_{+}}\sum_{i = 1}^{n_{+}}(y_{+, i} - x_{+, i}^\top \hat\beta)^2 \] where \(\hat\beta\) is the least squares estimator of the candidate model.
Choose the candidate model with the smallest \(\hat\Delta\).
Cross validation
Depending on \(n\), data splitting may not the most efficient use of the available information.
\(\Delta\) can instead be estimated using leave-one-out cross-validation as \[ \hat\Delta_\text{CV} = \frac{1}{n} \sum_{i=1}^n (y_i - x_i^\top \hat\beta_{-i})^2 \tag{4}\] where \(\hat\beta_{-j}\) is the estimate computed without the \(i\)th observation.
(4) suggests that we need \(n\) fits to compute \(\hat\Delta_\text{CV}\). But, it can be shown that \[ \hat\Delta_\text{CV} = \frac{1}{n} \sum_{i=1}^n \frac{(y_i - x_i^\top \hat\beta)^2}{(1 - h_{ii})^2} \] where \(h_{11},\ldots, h_{nn}\) are diagonal elements of \(H\).
Generalized cross validation
Another estimator of \(\Delta\) uses generalized cross-validation and has the form \[ \hat\Delta_\text{GCV} = \frac{1}{n} \sum_{i = 1}^n \frac{(y_i - x_i^\top \hat\beta)^2}{\{1 - \mathop{\mathrm{tr}}(H)/n\}^2} \]
It holds that \[ \mathop{\mathrm{E}}(\hat \Delta_\text{GCV}) = \frac{1}{n(1 - p / n)^2} \mu^\top (I_n - H)\mu + \frac{1}{1 - p / n} \sigma^2 \, . \] For large \(n\), \(\mathop{\mathrm{E}}(\hat \Delta_\text{GCV}) \approx \Delta\).
Generalized cross validation
Proof. It holds that \(Y - X\hat\beta = (I_n - H) Y = (I_n - H) \mu + (I_n - H) \epsilon\). So, \[ (Y - X\hat\beta)^\top (Y - X\hat\beta) = {\color[rgb]{1,1,1}{\mu^\top (I_n - H) \mu + \epsilon^\top (I_n - H) \epsilon + Z}} \] where the terms collected in \(Z\) have expectation zero. Using the fact that \(\mathop{\mathrm{tr}}{H} = p\), \[ \begin{aligned} \mathop{\mathrm{E}}(\hat \Delta_\text{GCV}) & = {\color[rgb]{1,1,1}{\frac{1}{n(1 - p / n)^2}\mu^\top (I_n - H) \mu + \frac{(n - p)}{n(1 - p / n)^2} \sigma^2}} \\ & = {\color[rgb]{1,1,1}{\frac{1}{n(1 - p / n)^2} \mu^\top (I_n - H) \mu + \frac{1}{1 - p/n}\sigma^2}} \end{aligned} \tag{5}\]
For large \(n\) or small \(z = 1 / n\), a Taylor expansion of \((1 - p z)^{-1}\) about \(z = 0\) gives \((1 - p z)^{-1} = {\color[rgb]{1,1,1}{1 + p z + O(z^2)}}\). Also, \(z (1 - z p)^{-2} = {\color[rgb]{1,1,1}{z + O(z^2)}}\). Replacing \((1 - p/n)^{-1}\) and \(n^{-1}(1 - p / n)^{-2}\) in the last expression in (5) with their first order approximations gives \[ \Delta = \mu (I_n - H) \mu / n + (1 + p / n) \sigma^2 \]
Bayesian model selection
Bayesian inference
Frequentist viewpoint (cartoon version)
There is a quantity \(\theta\) that characterizes aspects of how the data is generated and is treated as an unknown constant.
Probability statements concern randomness in hypothetical replications of the data (possibly conditioned on an ancillary statistic).
Bayesian viewpoint (cartoon version)
All ignorance should be expressed in terms of probability statements.
A joint probability distribution for data and all unknowns can be constructed.
Bayes’ theorem is used to convert prior beliefs \(\pi(\theta)\) about unknown \(\theta\) into posterior beliefs \(\pi(\theta\mid y)\), conditioned on data.
Probability statements concern randomness of unknowns, conditioned on all known quantities.

A joke by Xiao-Li Meng
Bayesian updating
Data \[ y \]
Likelihood (summarizes modelling assumptions / experimental conditions) \[ f(y \mid \theta), \quad \text{where} \quad \theta\in\Omega_\theta \]
Prior distribution (summarizes prior information about \(\theta\)) \[ \pi(\theta) \]
Posterior distribution (summarizes information about \(\theta\), conditional on the observed data \(y\)) \[ \pi(\theta\mid y ) = \frac{\pi(\theta) f(y\mid\theta)}{\int \pi(\theta) f(y\mid\theta)\, d\theta} \]
Marginal posterior
If \(\theta = (\psi^\top,\lambda^\top)^\top\), then inference for \(\psi\) is based on the marginal posterior \[ \pi(\psi\mid y) = \int \pi(\theta\mid y )\, d\lambda \]
Bayesian model selection
\(M\) alternative models for the data, with respective parameters \(\theta_1 \in \Omega_{\theta_1}, \ldots, \theta_M \in \Omega_{\theta_M}\)
Parameter space
\[ \theta \in \Omega = \bigcup_{m=1}^M \{m\}\times \Omega_{\theta_m} \]
Prior distributions for model parameters
\[ \pi_m(\theta_m \mid m) \]
Prior probabilities for models
\[ \pi(m) \]
Prior information for each model
\[ \pi(m, \theta_m) = \pi(\theta_m \mid m)\pi(m) \]
Inference about model choice
Marginal posterior probability mass function
\[ \begin{aligned} \pi(m\mid y) = \frac{\int f(y\mid\theta_m)\pi_m(\theta_m)\pi(m)\, d\theta_m} {\sum_{m'=1}^M \int f(y\mid\theta_{m'})\pi_{m'}(\theta_{m'})\pi(m')\, d\theta_{m'}} = \frac{\pi(m) \overbrace{f(y\mid m)}^{\text{marginal likelihood}}} {\sum_{m'=1}^M \pi(m') f(y\mid{m'})} \end{aligned} \]
Inference about models and parameters
Joint posterior of model and parameters
\[ \pi(m, \theta_m \mid y) =\pi(\theta_m \mid m, y)\pi(m \mid y) \]
Bayesian updating
\[ \pi(\theta_m\mid m)\pi(m) \mapsto \pi(\theta_m \mid m, y) \pi(m\mid y) \]
For each model \(m \in \{1,\ldots, M\}\), it is necessary to compute
\(\pi(m \mid y)\), which involves \(f(y\mid m) = \int f(y\mid\theta_m,m)\pi(\theta_m\mid m)\, d\theta_m\)
\(\pi(\theta_m \mid y, m)\)
Posterior odds = Prior odds \(\times\) Bayes factor
If there are just two models, we can write \[ \underbrace{\frac{\pi(1\mid y)}{\pi(2\mid y)}}_{\text{Posterior odds}} = \overbrace{\frac{\pi(1)}{\pi(2)}}^{\text{Prior odds}} \underbrace{\frac{f(y\mid 1)}{f(y\mid 2)}}_{\text{Bayes factor}} \]
Lindsey’s paradox
Suppose that \(\theta_m\) has as \(N(0, \sigma^2 I_{p_m})\) prior, where \(\sigma^2\) is large (prior is diffuse). Then, \[ \begin{aligned} f(y \mid m) &= \sigma^{-p_m} (2\pi)^{-p_m/2} \int f(y \mid m, \theta_m) \prod_{r = 1}^{p_m} \exp \left\{-{\theta_{m,r}^2/(2\sigma^2)}\right\} d\theta_{m, r}\\ & \approx \sigma^{-p_m} (2\pi)^{-p_m/2} \int f(y \mid m, \theta_m) \prod_{r = 1}^{p_m} d\theta_{m, r} \end{aligned} \]
The Bayes factor for comparing two models is \[ \frac{f(y\mid 1)}{f(y\mid 2)}\approx \sigma^{p_2 - p_1} g(y) \] where \(g(y)\) depends on the two likelihoods but is independent of \(\sigma^2\).
\[\LARGE\Downarrow\]
Irrespective of what the data tell us about the relative merits of the two models, the Bayes factor in favour of the simpler model can be made arbitrarily large by increasing \(\sigma\).
Model averaging
If a quantity \(Z\) has the same interpretation for all models, it is desirable to allow for model uncertainty when constructing inferences or making predictions about it.
Predictive distribution of \(Z\)
\[ f(z\mid y) = \sum_{m=1}^M f(z\mid m, y) \pi(m \mid y) \]
\(Z\) is a future value
Each model may be just a vehicle that provides a future value, and not of interest on its own.
\(Z\) is a physical parameter (e.g. means, variances, etc.)
Care is needed with prior choice and ensuring that \(Z\) has the same interpretation across models.
| Materials | Link |
|---|---|
| Preliminary material | ikosmidis.com/files/APTS-SM-Preliminary |
| Notes | ikosmidis.com/files/APTS-SM-Notes |
| Slides: Introduction | ikosmidis.com/files/APTS-SM-Slides-intro |
| Slides: Model selection | ikosmidis.com/files/APTS-SM-Slides-model-selection |
| Slides: Beyond GLMs | ikosmidis.com/files/APTS-SM-Slides-beyond-glms |
| Slides: Nonlinear models | ikosmidis.com/files/APTS-SM-Slides-nonlinear-models |
| Slides: Latent variables | ikosmidis.com/files/APTS-SM-Slides-latent |
| Lab 1 | ikosmidis.com/files/APTS-SM-Notes/lab1.html |
| Lab 2 | ikosmidis.com/files/APTS-SM-Notes/lab2.html |

Ioannis Kosmidis - Statistical Modelling: Model Selection

